Curate datasets 
 ¶
¶
Data curation with LaminDB ensures your datasets are validated and queryable through annotation.
Curating a dataset with LaminDB means three things:
Validate that the dataset matches a desired schema.
Standardize the dataset (e.g., by fixing typos, mapping synonyms) or update registries if validation fails.
Annotate the dataset by linking it against metadata entities so that it becomes queryable.
In this guide we’ll curate common data structures. Here is a guide for the underlying low-level API.
Note: If you know either pydantic or pandera, here is an FAQ that compares LaminDB with both of these tools.
# pip install lamindb
!lamin init --storage ./test-curate --modules bionty
Show code cell output
→ initialized lamindb: testuser1/test-curate
import lamindb as ln
ln.track()
Show code cell output
→ connected lamindb: testuser1/test-curate
→ created Transform('6oKxJGrdCjF50000', key='curate.ipynb'), started new Run('YxIrg2KglZVu7DDw') at 2026-03-06 16:23:28 UTC
→ notebook imports: lamindb==2.3a2
• recommendation: to identify the notebook across renames, pass the uid: ln.track("6oKxJGrdCjF5")
Schema design patterns¶
A Schema in LaminDB is a specification that defines the expected structure, data types, and validation rules for a dataset.
It is similar to pydantic.Model for dictionaries, and pandera.Schema, and pyarrow.lib.Schema for tables, but supporting more complicated data structures.
Schemas ensure data consistency by defining:
What
Features (dimensions) exist in your datasetWhat data types those features should have
What values are valid for categorical features
Which
Features are required vs optional
An exemplary schema:
schema = ln.Schema(
name="experiment_schema", # human-readable name
features=[ # required features
ln.Feature(name="cell_type", dtype=bt.CellType),
ln.Feature(name="treatment", dtype=str),
],
otype="DataFrame" # object type (DataFrame, AnnData, etc.)
)
For composite data structures using slots:
What are slots?
For composite data structures, you need to specify which component contains which schema, for example, to validate both cell metadata in .obs and gene metadata in .var within the same schema.
Each slot is a key like "obs" for AnnData observations,"rna:var" for MuData modalities, or "attrs:nested:key" for SpatialData annotations.
# AnnData with multiple "slots"
adata_schema = ln.Schema(
otype="AnnData",
slots={
"obs": cell_metadata_schema, # cell annotations
"var.T": gene_id_schema # gene-derived features
}
)
Before diving into curation, let’s understand the different schema approaches and when to use each one. Think of schemas as rules that define what valid data should look like.
Flexible schema¶
Use when: You want to validate those columns whose names match feature names in your Feature registry.
import lamindb as ln
schema = ln.Schema(name="valid_features", itype=ln.Feature).save()
Minimal required schema¶
Use when: You need certain columns but want flexibility for additional metadata.
import lamindb as ln
schema = ln.Schema(
name="Mini immuno schema",
features=[
ln.Feature.get(name="perturbation"),
ln.Feature.get(name="cell_type_by_model"),
ln.Feature.get(name="assay_oid"),
ln.Feature.get(name="donor"),
ln.Feature.get(name="concentration"),
ln.Feature.get(name="treatment_time_h"),
],
flexible=True, # _additional_ columns in a dataframe are validated & annotated
).save()
Strict Schema¶
Use when: You need complete control over data structure and values.
# Only allows specified columns
schema = ln.Schema(
features=[...],
minimal_set=True, # whether all passed features are required
maximal_set=False # whether additional features are allowed
)
DataFrame¶
Step 1: Load and examine your data¶
We’ll be working with the mini immuno dataset:
df = ln.examples.datasets.mini_immuno.get_dataset1(
with_cell_type_synonym=True, with_cell_type_typo=True
)
df
Show code cell output
| ENSG00000153563 | ENSG00000010610 | ENSG00000170458 | perturbation | sample_note | cell_type_by_expert | cell_type_by_model | assay_oid | concentration | treatment_time_h | donor | donor_ethnicity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sample1 | 1 | 3 | 5 | DMSO | was ok | B-cell | B cell | EFO:0008913 | 0.1% | 24 | D0001 | [Chinese, Singaporean Chinese] |
| sample2 | 2 | 4 | 6 | IFNG | looks naah | CD8-pos alpha-beta T cell | T cell | EFO:0008913 | 200 nM | 24 | D0002 | [Chinese, Han Chinese] |
| sample3 | 3 | 5 | 7 | DMSO | pretty! 🤩 | CD8-pos alpha-beta T cell | T cell | EFO:0008913 | 0.1% | 6 | None | [Chinese] |
Step 2: Set up your metadata registries¶
Before creating a schema, ensure your registries have the right features and labels:
import bionty as bt
import lamindb as ln
# define valid labels
perturbation_type = ln.Record(name="Perturbation", is_type=True).save()
ln.Record(name="DMSO", type=perturbation_type).save()
ln.Record(name="IFNG", type=perturbation_type).save()
bt.CellType.from_source(name="B cell").save()
bt.CellType.from_source(name="T cell").save()
# define valid features
ln.Feature(name="perturbation", dtype=perturbation_type).save()
ln.Feature(name="cell_type_by_expert", dtype=bt.CellType).save()
ln.Feature(name="cell_type_by_model", dtype=bt.CellType).save()
ln.Feature(name="assay_oid", dtype=bt.ExperimentalFactor.ontology_id).save()
ln.Feature(name="concentration", dtype=str).save()
ln.Feature(name="treatment_time_h", dtype="num", coerce=True).save()
ln.Feature(name="donor", dtype=str, nullable=True).save()
ln.Feature(name="donor_ethnicity", dtype=list[bt.Ethnicity]).save()
Step 3: Create your schema¶
schema = ln.examples.datasets.mini_immuno.define_mini_immuno_schema_flexible()
schema.describe()
Show code cell output
Schema: Mini immuno schema ├── uid: opk2CLbkVySOFw8e run: YxIrg2K (curate.ipynb) │ itype: Feature otype: None │ hash: eMTbY-rv4THgNwb_uYZQgw ordered_set: False │ maximal_set: False minimal_set: True │ branch: main space: all │ created_at: 2026-03-06 16:23:32 UTC created_by: testuser1 └── Features (6) └── name dtype optional nullable coerce default_value perturbation Record[Perturbation] ✗ ✓ ✗ unset cell_type_by_model bionty.CellType ✗ ✓ ✗ unset assay_oid bionty.ExperimentalFactor.ontology_id ✗ ✓ ✗ unset donor str ✗ ✓ ✗ unset concentration str ✗ ✓ ✗ unset treatment_time_h num ✗ ✓ ✓ unset
Step 4: Initialize Curator and first validation¶
If you expect the validation to pass, you can directly register an artifact by providing the schema:
artifact = ln.Artifact.from_dataframe(df, key="examples/my_curated_dataset.parquet", schema=schema).save()
The validate() method validates that your dataset adheres to the criteria defined by the schema.
It identifies which values are already validated (exist in the registries) and which are potentially problematic (do not yet exist in our registries).
try:
curator = ln.curators.DataFrameCurator(df, schema)
curator.validate()
except ln.errors.ValidationError as error:
print(error)
Show code cell output
! 4 terms not validated in feature 'columns': 'sample_note', 'ENSG00000170458', 'ENSG00000153563', 'ENSG00000010610'
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('columns')
! 2 terms not validated in feature 'cell_type_by_expert': 'B-cell', 'CD8-pos alpha-beta T cell'
1 synonym found: "B-cell" → "B cell"
→ curate synonyms via: .standardize("cell_type_by_expert")
for remaining terms:
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('cell_type_by_expert')
2 terms not validated in feature 'cell_type_by_expert': 'B-cell', 'CD8-pos alpha-beta T cell'
1 synonym found: "B-cell" → "B cell"
→ curate synonyms via: .standardize("cell_type_by_expert")
for remaining terms:
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('cell_type_by_expert')
Step 5: Fix validation issues¶
# check the non-validated terms
curator.cat.non_validated
Show code cell output
{'cell_type_by_expert': ['B-cell', 'CD8-pos alpha-beta T cell']}
For cell_type_by_expert, we saw 2 terms are not validated.
First, let’s standardize synonym “B-cell” as suggested
curator.cat.standardize("cell_type_by_expert")
# now we have only one non-validated cell type left
curator.cat.non_validated
Show code cell output
{'cell_type_by_expert': ['CD8-pos alpha-beta T cell']}
For “CD8-pos alpha-beta T cell”, let’s understand which cell type in the public ontology might be the actual match.
# to check the correct spelling of categories, pass `public=True` to get a lookup object from public ontologies
# use `lookup = curator.cat.lookup()` to get a lookup object of existing records in your instance
lookup = curator.cat.lookup(public=True)
lookup
Show code cell output
Lookup objects from the public:
.perturbation
.cell_type_by_expert
.cell_type_by_model
.assay_oid
.donor_ethnicity
.columns
Example:
→ categories = curator.lookup()["cell_type"]
→ categories.alveolar_type_1_fibroblast_cell
To look up public ontologies, use .lookup(public=True)
# here is an example for the "cell_type" column
cell_types = lookup["cell_type_by_expert"]
cell_types.cd8_positive_alpha_beta_t_cell
Show code cell output
CellType(ontology_id='CL:0000625', name='CD8-positive, alpha-beta T cell', definition='A T Cell Expressing An Alpha-Beta T Cell Receptor And The Cd8 Coreceptor.', synonyms='CD8-positive, alpha-beta T-cell|CD8-positive, alpha-beta T lymphocyte|CD8-positive, alpha-beta T-lymphocyte', parents=array(['CL:0000791'], dtype=object))
# fix the cell type name
df["cell_type_by_expert"] = df["cell_type_by_expert"].cat.rename_categories(
{"CD8-pos alpha-beta T cell": cell_types.cd8_positive_alpha_beta_t_cell.name}
)
For perturbation, we want to add the new values: “DMSO”, “IFNG”
# this adds perturbations that were _not_ validated
curator.cat.add_new_from("perturbation")
ln.Feature.get(name="perturbation")
Show code cell output
Feature(uid='CFGQ7dOgT5bK', is_type=False, name='perturbation', _dtype_str='cat[Record[rokdOAzzXFlHLKcf]]', unit=None, description=None, array_rank=0, array_size=0, array_shape=None, synonyms=None, default_value=None, nullable=True, coerce=None, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=1, type_id=None, created_at=2026-03-06 16:23:32 UTC, is_locked=False)
# validate again
curator.validate()
Show code cell output
! 4 terms not validated in feature 'columns': 'sample_note', 'ENSG00000170458', 'ENSG00000153563', 'ENSG00000010610'
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('columns')
Step 6: Save your curated dataset¶
artifact = curator.save_artifact(key="examples/my_curated_dataset.parquet")
artifact.describe()
Show code cell output
Artifact: examples/my_curated_dataset.parquet (0000) ├── uid: JOKrc7TGmQ7SXZch0000 run: YxIrg2K (curate.ipynb) │ kind: dataset otype: DataFrame │ hash: o2OEY7jSQiUOx4hxqrxaMw size: 10.1 KB │ branch: main space: all │ created_at: 2026-03-06 16:23:36 UTC created_by: testuser1 │ n_observations: 3 ├── storage/path: /home/runner/work/lamindb/lamindb/docs/test-curate/.lamindb/JOKrc7TGmQ7SXZch0000.parquet ├── Dataset features │ └── columns (8) │ assay_oid bionty.ExperimentalFactor.ontology… EFO:0008913 │ cell_type_by_expert bionty.CellType B cell, CD8-positive, alpha-beta T cell │ cell_type_by_model bionty.CellType B cell, T cell │ concentration str │ donor str │ donor_ethnicity list[bionty.Ethnicity] ['Chinese', 'Singaporean Chinese', 'Ha… │ perturbation Record[Perturbation] DMSO, IFNG │ treatment_time_h num └── Labels └── .records Record DMSO, IFNG .cell_types bionty.CellType B cell, T cell, CD8-positive, alpha-be… .experimental_factors bionty.ExperimentalFactor single-cell RNA sequencing .ethnicities bionty.Ethnicity Chinese, Singaporean Chinese, Han Chin…
Common fixes¶
This section covers the most frequent curation issues and their solutions. Use this as a reference when validation fails.
Feature validation issues¶
Issue: “Column not in dataframe”
"column 'treatment' not in dataframe. Columns in dataframe: ['drug', 'timepoint', ...]"
Solutions:
# Solution 1: Rename columns to match schema
df = df.rename(columns={
'treatment': 'drug',
'time': 'timepoint',
...
})
# Solution 2: Create missing columns
df['treatment'] = 'unknown' # Add with default value (or define Feature.default_value)
# Solution 3: Modify schema to match your data
schema = ln.Schema(
features=[
ln.Feature.get(name="drug"), # Use actual column name
ln.Feature.get(name="timepoint"),
],
...
)
Value validation issues¶
Issue: “Terms not validated in feature ‘perturbation’”
2 terms not validated in feature 'cell_type': 'B-cell', 'CD8-pos alpha-beta T cell'
1 synonym found: "B-cell" → "B cell"
→ curate synonyms via: .standardize("cell_type")
for remaining terms:
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('cell_type')
Solutions:
# Solution 1: Use automatic standardization if given hint (handles synonyms))
curator.cat.standardize('cell_type')
# Solution 2: Manual mapping for complex cases
value_mapping = {
'T-cells': 'T cell',
'B-cells': 'B cell',
}
df['cell_type'] = df['cell_type'].map(value_mapping).fillna(df['cell_type'])
# Solution 3: Use public ontology lookup for correct names
lookup = curator.cat.lookup(public=True)
cell_types = lookup["cell_type"]
df['cell_type'] = df['cell_type'].cat.rename_categories({
'CD8-pos T cell': cell_types.cd8_positive_alpha_beta_t_cell.name
})
# Solution 4: Add new legitimate terms
curator.cat.add_new_from("cell_type")
Data type issues¶
Issue: “Expected categorical data, got object”
TypeError: Expected categorical data for cell_type, got object
Solutions:
# Solution 1: Convert to categorical
df['cell_type'] = df['cell_type'].astype('category')
# Solution 2: Use coercion in feature definition
ln.Feature(name="cell_type", dtype=bt.CellType, coerce=True).save()
Organism-specific ontology issues¶
Issue: “Terms not validated” for organism-specific ontologies like developmental stages
2 terms not validated in feature 'developmental_stage_ontology_id': 'MmusDv:0000142', 'MmusDv:0000022'
Solution: Specify organism-specific source in feature definition using cat_filters:
# When defining the schema, specify the organism-specific source
mouse_source = bt.Source.filter(
entity="bionty.DevelopmentalStage",
organism="mouse"
).one()
schema = ln.Schema(
features=[
ln.Feature(
name="developmental_stage_ontology_id",
dtype=bt.DevelopmentalStage.ontology_id,
cat_filters={"source": mouse_source} # Specify organism-specific source
)
],
...
)
This pattern applies to any ontology where the same registry serves multiple organisms (e.g., DevelopmentalStage, Phenotype, …).
External data validation¶
Since not all metadata is always stored within the dataset itself, it is also possible to validate external metadata.
import lamindb as ln
from datetime import date
df = ln.examples.datasets.mini_immuno.get_dataset1(otype="DataFrame")
temperature = ln.Feature(name="temperature", dtype=float).save()
date_of_study = ln.Feature(name="date_of_study", dtype=date).save()
external_schema = ln.Schema(features=[temperature, date_of_study]).save()
concentration = ln.Feature(name="concentration", dtype=str).save()
donor = ln.Feature(name="donor", dtype=str, nullable=True).save()
schema = ln.Schema(
features=[concentration, donor],
slots={"__external__": external_schema},
otype="DataFrame",
).save()
artifact = ln.Artifact.from_dataframe(
df,
key="examples/dataset1.parquet",
features={"temperature": 21.6, "date_of_study": date(2024, 10, 1)},
schema=schema,
).save()
artifact.describe()
!python scripts/curate_dataframe_external_features.py
Show code cell output
→ connected lamindb: testuser1/test-curate
→ returning feature with same name: 'concentration'
→ returning feature with same name: 'donor'
! no run & transform got linked, call `ln.track()` & re-run
→ returning artifact with same hash: Artifact(uid='JOKrc7TGmQ7SXZch0000', version_tag=None, is_latest=True, key='examples/my_curated_dataset.parquet', description=None, suffix='.parquet', kind='dataset', otype='DataFrame', size=10354, hash='o2OEY7jSQiUOx4hxqrxaMw', n_files=None, n_observations=3, branch_id=1, created_on_id=1, space_id=1, storage_id=1, run_id=1, schema_id=1, created_by_id=1, created_at=2026-03-06 16:23:36 UTC, is_locked=False); to track this artifact as an input, use: ln.Artifact.get()
! key examples/my_curated_dataset.parquet on existing artifact differs from passed key examples/dataset1.parquet, keeping original key; update manually if needed or pass skip_hash_lookup if you want to duplicate the artifact
→ loading artifact into memory for validation
! 10 terms not validated in feature 'columns': 'treatment_time_h', 'ENSG00000153563', 'assay_oid', 'perturbation', 'sample_note', 'ENSG00000010610', 'cell_type_by_expert', 'cell_type_by_model', 'ENSG00000170458', 'donor_ethnicity'
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('columns')
Artifact: examples/my_curated_dataset.parquet (0000)
├── uid: JOKrc7TGmQ7SXZch0000 run: YxIrg2K (curate.ipynb)
│ kind: dataset otype: DataFrame
│ hash: o2OEY7jSQiUOx4hxqrxaMw size: 10.1 KB
│ branch: main space: all
│ created_at: 2026-03-06 16:23:36 UTC created_by: testuser1
│ n_observations: 3
├── storage/path:
│ /home/runner/work/lamindb/lamindb/docs/test-curate/.lamindb/JOKrc7TGmQ7SXZch
│ 0000.parquet
├── Dataset features
│ └── columns (2)
│ concentration str
│ donor str
├── External features
│ └── assay_oid bionty.ExperimentalFac… EFO:0008913
│ cell_type_by_expe… bionty.CellType B cell, CD8-positive, alph…
│ cell_type_by_model bionty.CellType B cell, T cell
│ donor_ethnicity list[bionty.Ethnicity] ['Chinese', 'Singaporean C…
│ perturbation Record[Perturbation] DMSO, IFNG
│ date_of_study date 2024-10-01
│ temperature float 21.6
└── Labels
└── .records Record DMSO, IFNG
.cell_types bionty.CellType B cell, T cell, CD8-positi…
.experimental_fac… bionty.ExperimentalFac… single-cell RNA sequencing
.ethnicities bionty.Ethnicity Chinese, Singaporean Chine…
Union dtypes¶
Some metadata columns might validate against several registries.
import lamindb as ln
import pandas as pd
union_feature = ln.Feature(
name="mixed_feature",
dtype="cat[bionty.Tissue.ontology_id|bionty.CellType.ontology_id]",
).save()
df_mixed = pd.DataFrame({"mixed_feature": ["UBERON:0000178", "CL:0000540"]})
schema = ln.Schema(features=[union_feature], coerce=True).save()
curator = ln.curators.DataFrameCurator(df_mixed, schema)
curator.validate()
!python scripts/curate_dataframe_union_features.py
Show code cell output
→ connected lamindb: testuser1/test-curate
! rather than passing a string 'cat[bionty.Tissue.ontology_id|bionty.CellType.ontology_id]' to dtype, consider passing a Python object
AnnData¶
AnnData like all other data structures that follow is a composite structure that stores different arrays in different slots.
Allow a flexible schema¶
We can also allow a flexible schema for an AnnData and only require that it’s indexed with Ensembl gene IDs.
import lamindb as ln
ln.examples.datasets.mini_immuno.define_features_labels()
adata = ln.examples.datasets.mini_immuno.get_dataset1(otype="AnnData")
artifact = ln.Artifact.from_anndata(
adata,
key="examples/mini_immuno.h5ad",
schema="ensembl_gene_ids_and_valid_features_in_obs",
).save()
artifact.describe()
Let’s run the script.
!python scripts/curate_anndata_flexible.py
Show code cell output
→ connected lamindb: testuser1/test-curate
→ returning record with same name: 'Perturbation'
→ returning record with same name: 'DMSO'
→ returning record with same name: 'IFNG'
→ returning feature with same name: 'perturbation'
→ returning feature with same name: 'cell_type_by_expert'
→ returning feature with same name: 'cell_type_by_model'
→ returning feature with same name: 'assay_oid'
→ returning feature with same name: 'concentration'
→ returning feature with same name: 'treatment_time_h'
→ returning feature with same name: 'donor'
→ returning feature with same name: 'donor_ethnicity'
! no run & transform got linked, call `ln.track()` & re-run
→ loading artifact into memory for validation
! 1 term not validated in feature 'columns' in slot 'obs': 'sample_note'
→ fix typos, remove non-existent values, or save terms via: curator.slots['obs'].cat.add_new_from('columns')
Artifact: examples/mini_immuno.h5ad (0000)
├── uid: ZLx1CzY5lpQuDb9v0000 run:
│ kind: dataset otype: AnnData
│ hash: FB3CeMjmg1ivN6HDy6wsSg size: 30.9 KB
│ branch: main space: all
│ created_at: 2026-03-06 16:23:52 UTC created_by: testuser1
│ n_observations: 3
├── storage/path:
│ /home/runner/work/lamindb/lamindb/docs/test-curate/.lamindb/ZLx1CzY5lpQuDb9v
│ 0000.h5ad
├── Dataset features
│ ├── obs (7)
│ │ assay_oid bionty.ExperimentalFac… EFO:0008913
│ │ cell_type_by_expe… bionty.CellType B cell, CD8-positive, alph…
│ │ cell_type_by_model bionty.CellType B cell, T cell
│ │ concentration str
│ │ donor str
│ │ perturbation Record[Perturbation] DMSO, IFNG
│ │ treatment_time_h num
│ └── var.T (3 bionty.G…
│ CD14 num
│ CD4 num
│ CD8A num
└── Labels
└── .records Record DMSO, IFNG
.cell_types bionty.CellType B cell, T cell, CD8-positi…
.experimental_fac… bionty.ExperimentalFac… single-cell RNA sequencing
Under-the-hood, this uses the following build-in schema (anndata_ensembl_gene_ids_and_valid_features_in_obs()):
import bionty as bt
import lamindb as ln
obs_schema = ln.examples.schemas.valid_features()
varT_schema = ln.Schema(
name="valid_ensembl_gene_ids", itype=bt.Gene.ensembl_gene_id
).save()
schema = ln.Schema(
name="anndata_ensembl_gene_ids_and_valid_features_in_obs",
otype="AnnData",
slots={"obs": obs_schema, "var.T": varT_schema},
).save()
This schema tranposes the var DataFrame during curation, so that one validates and annotates the columns of var.T, i.e., [ENSG00000153563, ENSG00000010610, ENSG00000170458].
If one doesn’t transpose, one would annotate the columns of var, i.e., [gene_symbol, gene_type].
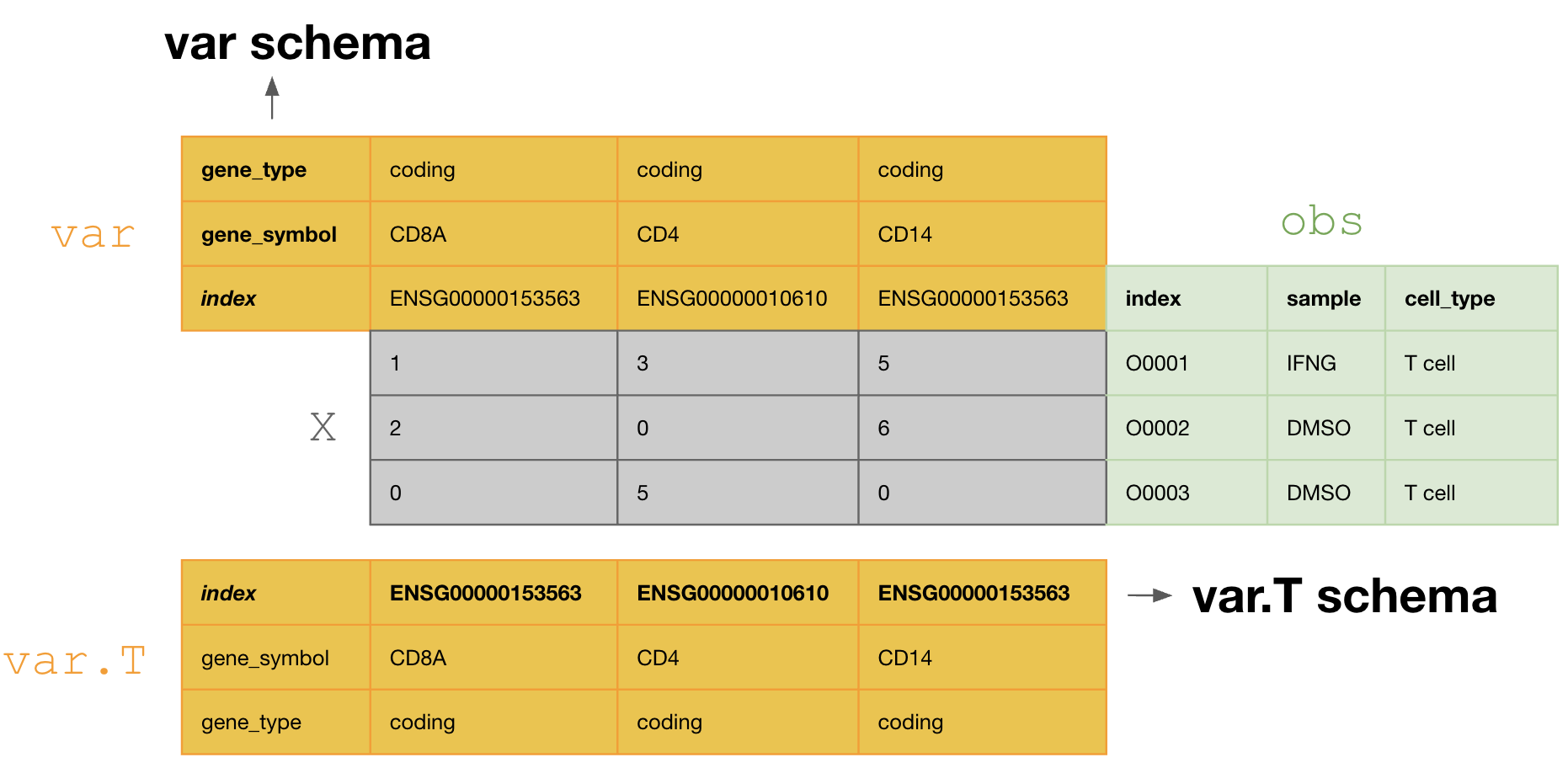
Fix validation issues¶
adata = ln.examples.datasets.mini_immuno.get_dataset1(
with_gene_typo=True, with_cell_type_typo=True, otype="AnnData"
)
adata
Show code cell output
AnnData object with n_obs × n_vars = 3 × 3
obs: 'perturbation', 'sample_note', 'cell_type_by_expert', 'cell_type_by_model', 'assay_oid', 'concentration', 'treatment_time_h', 'donor'
uns: 'temperature', 'experiment', 'date_of_study', 'study_note'
schema = ln.examples.schemas.anndata_ensembl_gene_ids_and_valid_features_in_obs()
schema.describe()
Show code cell output
Schema: anndata_ensembl_gene_ids_and_valid_features_in_obs ├── uid: 0000000000000002 run: │ itype: None otype: AnnData │ hash: aqGWHvyY49W_PHELUMiBMw ordered_set: False │ maximal_set: False minimal_set: True │ branch: main space: all │ created_at: 2026-03-06 16:23:48 UTC created_by: testuser1 ├── obs: valid_features │ └── uid: 0000000000000000 run: │ itype: Feature otype: None │ hash: kMi7B_N88uu-YnbTLDU-DA ordered_set: False │ maximal_set: False minimal_set: True │ branch: main space: all │ created_at: 2026-03-06 16:23:48 UTC created_by: testuser1 └── var.T: valid_ensembl_gene_ids ├── uid: 0000000000000001 run: │ itype: bionty.Gene.ensembl_gene_id otype: None │ hash: 1gocc_TJ1RU2bMwDRK-WUA ordered_set: False │ maximal_set: False minimal_set: True │ branch: main space: all │ created_at: 2026-03-06 16:23:48 UTC created_by: testuser1 └── bionty.Gene.ensembl_gene_id └── dtype: num
Check the slots of a schema:
schema.slots
Show code cell output
{'obs': Schema(uid='0000000000000000', is_type=False, name='valid_features', description=None, n_members=None, coerce=None, flexible=True, itype='Feature', otype=None, hash='kMi7B_N88uu-YnbTLDU-DA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=None, type_id=None, created_at=2026-03-06 16:23:48 UTC, is_locked=False),
'var.T': Schema(uid='0000000000000001', is_type=False, name='valid_ensembl_gene_ids', description=None, n_members=None, coerce=None, flexible=True, itype='bionty.Gene.ensembl_gene_id', otype=None, hash='1gocc_TJ1RU2bMwDRK-WUA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=None, type_id=None, created_at=2026-03-06 16:23:48 UTC, is_locked=False)}
curator = ln.curators.AnnDataCurator(adata, schema)
try:
curator.validate()
except ln.errors.ValidationError as error:
print(error)
Show code cell output
! 1 term not validated in feature 'columns' in slot 'obs': 'sample_note'
→ fix typos, remove non-existent values, or save terms via: curator.slots['obs'].cat.add_new_from('columns')
! 1 term not validated in feature 'cell_type_by_expert' in slot 'obs': 'CD8-pos alpha-beta T cell'
→ fix typos, remove non-existent values, or save terms via: curator.slots['obs'].cat.add_new_from('cell_type_by_expert')
1 term not validated in feature 'cell_type_by_expert' in slot 'obs': 'CD8-pos alpha-beta T cell'
→ fix typos, remove non-existent values, or save terms via: curator.slots['obs'].cat.add_new_from('cell_type_by_expert')
As above, we leverage a lookup object with valid cell types to find the correct name.
valid_cell_types = curator.slots["obs"].cat.lookup()["cell_type_by_expert"]
adata.obs["cell_type_by_expert"] = adata.obs[
"cell_type_by_expert"
].cat.rename_categories(
{"CD8-pos alpha-beta T cell": valid_cell_types.cd8_positive_alpha_beta_t_cell.name}
)
The validated AnnData can be subsequently saved as an Artifact:
adata.obs.columns
Show code cell output
Index(['perturbation', 'sample_note', 'cell_type_by_expert',
'cell_type_by_model', 'assay_oid', 'concentration', 'treatment_time_h',
'donor'],
dtype='object')
curator.slots["var.T"].cat.add_new_from("columns")
Show code cell output
! 1 term not validated in feature 'columns' in slot 'var.T': 'GeneTypo'
→ fix typos, remove non-existent values, or save terms via: curator.slots['var.T'].cat.add_new_from('columns')
curator.validate()
Show code cell output
! 1 term not validated in feature 'columns' in slot 'obs': 'sample_note'
→ fix typos, remove non-existent values, or save terms via: curator.slots['obs'].cat.add_new_from('columns')
artifact = curator.save_artifact(key="examples/my_curated_anndata.h5ad")
Show code cell output
→ returning schema with same hash: Schema(uid='7JVaWeDDn9iie1xn', is_type=False, name=None, description=None, n_members=7, coerce=None, flexible=False, itype='Feature', otype=None, hash='SrkH6_UbG3wZXWqrrCmJaA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=None, type_id=None, created_at=2026-03-06 16:23:52 UTC, is_locked=False)
! run was not set on Schema(uid='7JVaWeDDn9iie1xn', is_type=False, name=None, description=None, n_members=7, coerce=None, flexible=False, itype='Feature', otype=None, hash='SrkH6_UbG3wZXWqrrCmJaA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=None, type_id=None, created_at=2026-03-06 16:23:52 UTC, is_locked=False), setting to current run
Access the schema for each slot:
artifact.features.slots
Show code cell output
{'obs': Schema(uid='7JVaWeDDn9iie1xn', is_type=False, name=None, description=None, n_members=7, coerce=None, flexible=False, itype='Feature', otype=None, hash='SrkH6_UbG3wZXWqrrCmJaA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=1, type_id=None, created_at=2026-03-06 16:23:52 UTC, is_locked=False),
'var.T': Schema(uid='BxkR62LZDcUS6Poi', is_type=False, name=None, description=None, n_members=3, coerce=None, flexible=False, itype='bionty.Gene.ensembl_gene_id', otype=None, hash='t9Nx3R1yIOM-v6okdPFsiw', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=1, type_id=None, created_at=2026-03-06 16:23:57 UTC, is_locked=False)}
The saved artifact has been annotated with validated features and labels:
artifact.describe()
Show code cell output
Artifact: examples/my_curated_anndata.h5ad (0000) ├── uid: 3NaMZWYaRjiI28T20000 run: YxIrg2K (curate.ipynb) │ kind: dataset otype: AnnData │ hash: yeNWx0-dOGGkANQbocU4Sg size: 30.9 KB │ branch: main space: all │ created_at: 2026-03-06 16:23:56 UTC created_by: testuser1 │ n_observations: 3 ├── storage/path: /home/runner/work/lamindb/lamindb/docs/test-curate/.lamindb/3NaMZWYaRjiI28T20000.h5ad ├── Dataset features │ ├── obs (7) │ │ assay_oid bionty.ExperimentalFactor.ontology… EFO:0008913 │ │ cell_type_by_expert bionty.CellType B cell, CD8-positive, alpha-beta T cell │ │ cell_type_by_model bionty.CellType B cell, T cell │ │ concentration str │ │ donor str │ │ perturbation Record[Perturbation] DMSO, IFNG │ │ treatment_time_h num │ └── var.T (3 bionty.Gene.ensembl… │ CD4 num │ CD8A num └── Labels └── .records Record DMSO, IFNG .cell_types bionty.CellType B cell, T cell, CD8-positive, alpha-be… .experimental_factors bionty.ExperimentalFactor single-cell RNA sequencing
Unstructured dictionaries¶
Most datastructures support unstructured metadata stored as dictionaries:
Pandas DataFrames:
.attrsAnnData:
.unsMuData:
.unsandmodality:unsSpatialData:
.attrs
Here, we exemplary show how to curate such metadata for AnnData:
import lamindb as ln
from define_schema_df_metadata import study_metadata_schema
anndata_uns_schema = ln.Schema(
otype="AnnData",
slots={
"uns:study_metadata": study_metadata_schema,
},
).save()
!python scripts/define_schema_anndata_uns.py
Show code cell output
→ connected lamindb: testuser1/test-curate
→ returning feature with same name: 'temperature'
import lamindb as ln
ln.examples.datasets.mini_immuno.define_features_labels()
adata = ln.examples.datasets.mini_immuno.get_dataset1(otype="AnnData")
schema = ln.Schema.get(name="Study metadata schema")
artifact = ln.Artifact.from_anndata(
adata, schema=schema, key="examples/mini_immuno_uns.h5ad"
)
artifact.describe()
!python scripts/curate_anndata_uns.py
Show code cell output
→ connected lamindb: testuser1/test-curate
→ returning record with same name: 'Perturbation'
→ returning record with same name: 'DMSO'
→ returning record with same name: 'IFNG'
→ returning feature with same name: 'perturbation'
→ returning feature with same name: 'cell_type_by_expert'
→ returning feature with same name: 'cell_type_by_model'
→ returning feature with same name: 'assay_oid'
→ returning feature with same name: 'concentration'
→ returning feature with same name: 'treatment_time_h'
→ returning feature with same name: 'donor'
→ returning feature with same name: 'donor_ethnicity'
! no run & transform got linked, call `ln.track()` & re-run
→ returning artifact with same hash: Artifact(uid='ZLx1CzY5lpQuDb9v0000', version_tag=None, is_latest=True, key='examples/mini_immuno.h5ad', description=None, suffix='.h5ad', kind='dataset', otype='AnnData', size=31672, hash='FB3CeMjmg1ivN6HDy6wsSg', n_files=None, n_observations=3, branch_id=1, created_on_id=1, space_id=1, storage_id=1, run_id=None, schema_id=9, created_by_id=1, created_at=2026-03-06 16:23:52 UTC, is_locked=False); to track this artifact as an input, use: ln.Artifact.get()
! key examples/mini_immuno.h5ad on existing artifact differs from passed key examples/mini_immuno_uns.h5ad, keeping original key; update manually if needed or pass skip_hash_lookup if you want to duplicate the artifact
→ loading artifact into memory for validation
Traceback (most recent call last):
File "/home/runner/work/lamindb/lamindb/docs/scripts/curate_anndata_uns.py", line 6, in <module>
artifact = ln.Artifact.from_anndata(
^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/runner/work/lamindb/lamindb/lamindb/models/artifact.py", line 2210, in from_anndata
curator = AnnDataCurator(artifact, schema)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/runner/work/lamindb/lamindb/lamindb/curators/core.py", line 1003, in __init__
raise InvalidArgument("Schema otype must be 'AnnData'.")
lamindb.errors.InvalidArgument: Schema otype must be 'AnnData'.
MuData¶
import lamindb as ln
import bionty as bt
from docs.scripts.define_schema_df_metadata import study_metadata_schema
# define labels
perturbation = ln.Record(name="Perturbation", is_type=True).save()
ln.Record(name="Perturbed", type=perturbation).save()
ln.Record(name="NT", type=perturbation).save()
replicate = ln.Record(name="Replicate", is_type=True).save()
ln.Record(name="rep1", type=replicate).save()
ln.Record(name="rep2", type=replicate).save()
ln.Record(name="rep3", type=replicate).save()
# define the global obs schema
obs_schema = ln.Schema(
name="mudata_papalexi21_subset_obs_schema",
features=[
ln.Feature(name="perturbation", dtype="cat[Record[Perturbation]]").save(),
ln.Feature(name="replicate", dtype="cat[Record[Replicate]]").save(),
],
).save()
# define the ['rna'].obs schema
obs_schema_rna = ln.Schema(
name="mudata_papalexi21_subset_rna_obs_schema",
features=[
ln.Feature(name="nCount_RNA", dtype=int).save(),
ln.Feature(name="nFeature_RNA", dtype=int).save(),
ln.Feature(name="percent.mito", dtype=float).save(),
],
).save()
# define the ['hto'].obs schema
obs_schema_hto = ln.Schema(
name="mudata_papalexi21_subset_hto_obs_schema",
features=[
ln.Feature(name="nCount_HTO", dtype=float).save(),
ln.Feature(name="nFeature_HTO", dtype=int).save(),
ln.Feature(name="technique", dtype=bt.ExperimentalFactor).save(),
],
).save()
# define ['rna'].var schema
var_schema_rna = ln.Schema(
name="mudata_papalexi21_subset_rna_var_schema",
itype=bt.Gene.symbol,
dtype=float,
).save()
# define composite schema
mudata_schema = ln.Schema(
name="mudata_papalexi21_subset_mudata_schema",
otype="MuData",
slots={
"obs": obs_schema,
"rna:obs": obs_schema_rna,
"hto:obs": obs_schema_hto,
"rna:var": var_schema_rna,
"uns:study_metadata": study_metadata_schema,
},
).save()
# curate a MuData
mdata = ln.examples.datasets.mudata_papalexi21_subset(with_uns=True)
bt.settings.organism = "human" # set the organism to map gene symbols
curator = ln.curators.MuDataCurator(mdata, mudata_schema)
artifact = curator.save_artifact(key="examples/mudata_papalexi21_subset.h5mu")
assert artifact.schema == mudata_schema
!python scripts/curate_mudata.py
Show code cell output
→ connected lamindb: testuser1/test-curate
→ returning feature with same name: 'temperature'
→ returning feature with same name: 'experiment'
→ returning schema with same hash: Schema(uid='nKmZmrcvrdIAK8Jz', is_type=False, name='Study metadata schema', description=None, n_members=2, coerce=None, flexible=False, itype='Feature', otype=None, hash='icv_n5uk4GnEBx7mSC48IQ', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=None, type_id=None, created_at=2026-03-06 16:23:58 UTC, is_locked=False)
→ returning record with same name: 'Perturbation'
! rather than passing a string 'cat[Record[Perturbation]]' to dtype, consider passing a Python object
→ returning feature with same name: 'perturbation'
! rather than passing a string 'cat[Record[Replicate]]' to dtype, consider passing a Python object
! you are trying to create a record with name='nFeature_HTO' but a record with similar name exists: 'nFeature_RNA'. Did you mean to load it?
/opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/mudata/_core/mudata.py:1783: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
mod_df.rename(
! auto-transposed `var` for backward compat, please indicate transposition in the schema definition by calling out `.T`: slots={'var.T': itype=bt.Gene.ensembl_gene_id}
! 37 terms not validated in feature 'columns' in slot 'obs': 'adt:S.Score', 'adt:NT', 'hto:MULTI_ID', 'adt:guide_ID', 'gdo:HTO_classification', 'adt:HTO_classification', 'hto:Phase', 'gdo:NT', 'hto:S.Score', 'gdo:orig.ident', 'gdo:guide_ID', 'hto:replicate', 'gdo:S.Score', 'hto:NT', 'gdo:G2M.Score', 'hto:guide_ID', 'adt:perturbation', 'hto:HTO_classification', 'adt:gene_target', 'gdo:MULTI_ID', ...
→ fix typos, remove non-existent values, or save terms via: curator.slots['obs'].cat.add_new_from('columns')
! 96 terms not validated in feature 'columns' in slot 'rna:var': 'RP5-827C21.6', 'XX-CR54.1', 'RP11-379B18.5', 'RP11-778D9.12', 'RP11-703G6.1', 'AC005150.1', 'RP11-717H13.1', 'CTC-498J12.1', 'CTC-467M3.1', 'HIST1H4K', 'RP11-524H19.2', 'AC006042.7', 'AC002066.1', 'AC073934.6', 'RP11-268G12.1', 'U52111.14', 'RP11-235C23.5', 'RP11-12J10.3', 'CASC1', 'RP11-324E6.9', ...
12 synonyms found: "CTC-467M3.1" → "MEF2C-AS2", "HIST1H4K" → "H4C12", "CASC1" → "DNAI7", "LARGE" → "LARGE1", "NBPF16" → "NBPF15", "C1orf65" → "CCDC185", "IBA57-AS1" → "IBA57-DT", "KIAA1239" → "NWD2", "TMEM75" → "LINC02912", "AP003419.16" → "RPS6KB2-AS1", "FAM65C" → "RIPOR3", "C14orf177" → "LINC02914"
→ curate synonyms via: .standardize("columns")
for remaining terms:
→ fix organism 'Organism(uid='1dpCL6TduFJ3AP', name='human', ontology_id='NCBITaxon:9606', abbr=None, synonyms=None, description=None, scientific_name='Homo sapiens', branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=None, source_id=1, created_at=2026-03-06 16:23:49 UTC, is_locked=False)', fix typos, remove non-existent values, or save terms via: curator.slots['rna:var'].cat.add_new_from('columns')
! no run & transform got linked, call `ln.track()` & re-run
→ returning schema with same hash: Schema(uid='odalKxClTt03FtTh', is_type=False, name='mudata_papalexi21_subset_obs_schema', description=None, n_members=2, coerce=None, flexible=False, itype='Feature', otype=None, hash='cDu7eHuevpyGSzsz7WswiQ', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=None, type_id=None, created_at=2026-03-06 16:24:05 UTC, is_locked=False)
→ returning schema with same hash: Schema(uid='bQKHlcJOsFPzNkXL', is_type=False, name='mudata_papalexi21_subset_rna_obs_schema', description=None, n_members=3, coerce=None, flexible=False, itype='Feature', otype=None, hash='EvIxV1SW0UmhQGkL_XceOA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=None, type_id=None, created_at=2026-03-06 16:24:05 UTC, is_locked=False)
→ returning schema with same hash: Schema(uid='FXBHBOhRgqOh7ZiX', is_type=False, name='mudata_papalexi21_subset_hto_obs_schema', description=None, n_members=3, coerce=None, flexible=False, itype='Feature', otype=None, hash='jo-amVwNVQv4CEoiUcVVXQ', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=None, type_id=None, created_at=2026-03-06 16:24:05 UTC, is_locked=False)
→ returning schema with same hash: Schema(uid='nKmZmrcvrdIAK8Jz', is_type=False, name='Study metadata schema', description=None, n_members=2, coerce=None, flexible=False, itype='Feature', otype=None, hash='icv_n5uk4GnEBx7mSC48IQ', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=None, type_id=None, created_at=2026-03-06 16:23:58 UTC, is_locked=False)
SpatialData¶
import lamindb as ln
import bionty as bt
attrs_schema = ln.Schema(
features=[
ln.Feature(name="bio", dtype=dict).save(),
ln.Feature(name="tech", dtype=dict).save(),
],
).save()
sample_schema = ln.Schema(
features=[
ln.Feature(name="disease", dtype=bt.Disease, coerce=True).save(),
ln.Feature(
name="developmental_stage",
dtype=bt.DevelopmentalStage,
coerce=True,
).save(),
],
).save()
tech_schema = ln.Schema(
features=[
ln.Feature(name="assay", dtype=bt.ExperimentalFactor, coerce=True).save(),
],
).save()
obs_schema = ln.Schema(
features=[
ln.Feature(name="sample_region", dtype="str").save(),
],
).save()
uns_schema = ln.Schema(
features=[
ln.Feature(name="analysis", dtype="str").save(),
],
).save()
# Schema enforces only registered Ensembl Gene IDs are valid (maximal_set=True)
varT_schema = ln.Schema(itype=bt.Gene.ensembl_gene_id, maximal_set=True).save()
sdata_schema = ln.Schema(
name="spatialdata_blobs_schema",
otype="SpatialData",
slots={
"attrs:bio": sample_schema,
"attrs:tech": tech_schema,
"attrs": attrs_schema,
"tables:table:obs": obs_schema,
"tables:table:var.T": varT_schema,
},
).save()
!python scripts/define_schema_spatialdata.py
Show code cell output
→ connected lamindb: testuser1/test-curate
! you are trying to create a record with name='tech' but a record with similar name exists: 'technique'. Did you mean to load it?
! you are trying to create a record with name='assay' but a record with similar name exists: 'assay_oid'. Did you mean to load it?
! rather than passing a string 'str' to dtype, consider passing a Python object
! rather than passing a string 'str' to dtype, consider passing a Python object
import lamindb as ln
spatialdata = ln.examples.datasets.spatialdata_blobs()
sdata_schema = ln.Schema.get(name="spatialdata_blobs_schema")
curator = ln.curators.SpatialDataCurator(spatialdata, sdata_schema)
try:
curator.validate()
except ln.errors.ValidationError:
pass
spatialdata.tables["table"].var.drop(index="ENSG00000999999", inplace=True)
# validate again (must pass now) and save artifact
artifact = ln.Artifact.from_spatialdata(
spatialdata, key="examples/spatialdata1.zarr", schema=sdata_schema
).save()
artifact.describe()
!python scripts/curate_spatialdata.py
Show code cell output
→ connected lamindb: testuser1/test-curate
Traceback (most recent call last):
File "/home/runner/work/lamindb/lamindb/docs/scripts/curate_spatialdata.py", line 3, in <module>
spatialdata = ln.examples.datasets.spatialdata_blobs()
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/runner/work/lamindb/lamindb/lamindb/examples/datasets/_core.py", line 607, in spatialdata_blobs
from spatialdata.datasets import blobs
File "/opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/spatialdata/__init__.py", line 72, in <module>
from spatialdata._io._utils import get_dask_backing_files
File "/opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/spatialdata/_io/__init__.py", line 1, in <module>
from spatialdata._io._utils import get_dask_backing_files
File "/opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/spatialdata/_io/_utils.py", line 24, in <module>
from zarr.storage import FsspecStore, LocalStore
ImportError: cannot import name 'FsspecStore' from 'zarr.storage' (/opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/zarr/storage.py)
TiledbsomaExperiment¶
import lamindb as ln
import bionty as bt
import tiledbsoma as soma
import tiledbsoma.io
adata = ln.examples.datasets.mini_immuno.get_dataset1(otype="AnnData")
tiledbsoma.io.from_anndata("small_dataset.tiledbsoma", adata, measurement_name="RNA")
obs_schema = ln.Schema(
name="soma_obs_schema",
features=[
ln.Feature(name="cell_type_by_expert", dtype=bt.CellType).save(),
ln.Feature(name="cell_type_by_model", dtype=bt.CellType).save(),
],
).save()
var_schema = ln.Schema(
name="soma_var_schema",
features=[
ln.Feature(name="var_id", dtype=bt.Gene.ensembl_gene_id).save(),
],
coerce=True,
).save()
soma_schema = ln.Schema(
name="soma_experiment_schema",
otype="tiledbsoma",
slots={
"obs": obs_schema,
"ms:RNA.T": var_schema,
},
).save()
with soma.Experiment.open("small_dataset.tiledbsoma") as experiment:
curator = ln.curators.TiledbsomaExperimentCurator(experiment, soma_schema)
curator.validate()
artifact = curator.save_artifact(
key="examples/soma_experiment.tiledbsoma",
description="SOMA experiment with schema validation",
)
assert artifact.schema == soma_schema
artifact.describe()
!python scripts/curate_soma_experiment.py
Show code cell output
→ connected lamindb: testuser1/test-curate
→ returning feature with same name: 'cell_type_by_expert'
→ returning feature with same name: 'cell_type_by_model'
! 6 terms not validated in feature 'columns': 'sample_note', 'perturbation', 'concentration', 'treatment_time_h', 'donor', 'assay_oid'
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('columns')
! no run & transform got linked, call `ln.track()` & re-run
→ returning schema with same hash: Schema(uid='VA97ckeAJFd3GPzK', is_type=False, name='soma_obs_schema', description=None, n_members=2, coerce=None, flexible=False, itype='Feature', otype=None, hash='fbciPKwM3tiGhvteChGEhQ', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=None, type_id=None, created_at=2026-03-06 16:24:24 UTC, is_locked=False)
→ returning schema with same hash: Schema(uid='VhOdezxDhqNe9X84', is_type=False, name='soma_var_schema', description=None, n_members=1, coerce=True, flexible=False, itype='Feature', otype=None, hash='vz3lvbXprRjO0aFIlTg17g', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=None, type_id=None, created_at=2026-03-06 16:24:24 UTC, is_locked=False)
Artifact: examples/soma_experiment.tiledbsoma (0000)
| description: SOMA experiment with schema validation
├── uid: hyEimbtrFpKgE6Px0000 run:
│ kind: dataset otype: tiledbsoma
│ hash: 8-LnTEatQzk4qWFv2_Y3bA size: 23.9 KB
│ branch: main space: all
│ created_at: 2026-03-06 16:24:25 UTC created_by: testuser1
│ n_files: 68 n_observations: 3
├── storage/path:
│ /home/runner/work/lamindb/lamindb/docs/test-curate/.lamindb/hyEimbtrFpKgE6Px
│ .tiledbsoma
├── Dataset features
│ ├── obs (2)
│ │ cell_type_by_expe… bionty.CellType B cell, CD8-positive, alph…
│ │ cell_type_by_model bionty.CellType B cell, T cell
│ └── ms:RNA.T (1)
│ var_id bionty.Gene.ensembl_ge… ENSG00000010610, ENSG00000…
└── Labels
└── .genes bionty.Gene CD8A, CD4, CD14
.cell_types bionty.CellType B cell, T cell, CD8-positi…
Other data structures¶
If you have other data structures, read: How do I validate & annotate arbitrary data structures? .
!rm -rf ./test-curate
!rm -rf ./small_dataset.tiledbsoma
!lamin delete --force test-curate