Redun 
 ¶
¶
Here, we’ll see how to track redun workflows with lamindb.
Note
This guide is based on github.com/ricomnl/bioinformatics-pipeline-tutorial.
Amend the workflow¶
Here is how to instrument a redun workflow for tracking with lamindb:
Add
ln.track()to themain()task (see on GitHub)Register desired output files or folders by creating artifacts for them (see on GitHub):
ln.Artifact(output_path, key="data/results.tar.gz").save()
Add a
finish()task that callsln.finish()(see on GitHub)Optionally cache/stage input files (see on GitHub)
Why not use @ln.flow() for main()?
Because main() in redun is typically a scheduler/executor task rather than a task that performs the actual computation. ln.flow() would then just track the execution time of scheduling, and not an actual compute run.
If one wanted to use @ln.flow() it’s advisable to wrap the scheduling main() task:
@ln.flow()
def run_pipeline(...):
scheduler = Scheduler()
result = scheduler.run(main(...)) # run the main task
ln.Artifact(result.path, key="data/results.tgz").save()
return result
Run redun¶
Let’s see what the input files are:
!ls ./fasta
Show code cell output
KLF4.fasta MYC.fasta PO5F1.fasta SOX2.fasta
Create a lamindb test instance:
# pip install lamindb redun git+http://github.com/laminlabs/redun-lamin-fasta
!lamin init --storage ./test-redun-lamin
Show code cell output
→ initialized lamindb: testuser1/test-redun-lamin
Register each input file individually as an artifact:
import lamindb as ln
import json
ln.Artifact.from_dir("./fasta").save()
Show code cell output
→ connected lamindb: testuser1/test-redun-lamin
! folder is outside existing storage location, will copy files from ./fasta to /home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/fasta
! no run & transform got linked, call `ln.track()` & re-run
! no run & transform got linked, call `ln.track()` & re-run
! no run & transform got linked, call `ln.track()` & re-run
! no run & transform got linked, call `ln.track()` & re-run
SQLRecordList([Artifact(uid='c44ljlhVZeSLf1W30000', version_tag=None, is_latest=True, key='fasta/SOX2.fasta', description=None, suffix='.fasta', kind=None, otype=None, size=414, hash='C5q_yaFXGk4SAEpfdqBwnQ', n_files=None, n_observations=None, branch_id=1, created_on_id=1, space_id=1, storage_id=1, run_id=None, schema_id=None, created_by_id=1, created_at=2026-03-06 16:22:49 UTC, is_locked=False),
Artifact(uid='bXtyUfEfXnr080JI0000', version_tag=None, is_latest=True, key='fasta/MYC.fasta', description=None, suffix='.fasta', kind=None, otype=None, size=536, hash='WGbEtzPw-3bQEGcngO_pHQ', n_files=None, n_observations=None, branch_id=1, created_on_id=1, space_id=1, storage_id=1, run_id=None, schema_id=None, created_by_id=1, created_at=2026-03-06 16:22:49 UTC, is_locked=False),
Artifact(uid='QoNT2toucir3QJWw0000', version_tag=None, is_latest=True, key='fasta/PO5F1.fasta', description=None, suffix='.fasta', kind=None, otype=None, size=477, hash='-7iJgveFO9ia0wE1bqVu6g', n_files=None, n_observations=None, branch_id=1, created_on_id=1, space_id=1, storage_id=1, run_id=None, schema_id=None, created_by_id=1, created_at=2026-03-06 16:22:49 UTC, is_locked=False),
Artifact(uid='CtiEqWncr9uDvcbr0000', version_tag=None, is_latest=True, key='fasta/KLF4.fasta', description=None, suffix='.fasta', kind=None, otype=None, size=609, hash='LyuoYkWs4SgYcH7P7JLJtA', n_files=None, n_observations=None, branch_id=1, created_on_id=1, space_id=1, storage_id=1, run_id=None, schema_id=None, created_by_id=1, created_at=2026-03-06 16:22:49 UTC, is_locked=False)])
Run the redun workflow:
!redun run workflow.py main --input-dir ./fasta --tag run=test-run 1> run_logs.txt 2>run_logs.txt
Inspect the logs:
!cat run_logs.txt
Show code cell output
→ connected lamindb: testuser1/test-redun-lamin
→ script invoked with: run workflow.py main --input-dir ./fasta --tag run=test-run
→ created Transform('75jlbxVuCz0i0000', key='workflow.py'), started new Run('Bz2tietYvi0bBPtS') at 2026-03-06 16:22:58 UTC
• recommendation: to identify the script across renames, pass the uid: ln.track("75jlbxVuCz0i")
File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/results.tgz, hash=4ad34654)
_tutorial.lib.digest_protein_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/c44ljlhVZeSLf1W30000.fasta, hash=298f7f94), enzyme_regex='[KR]', missed_cleavages=0, min_length=4, max_length=75) on default
[redun] Run Job 4caa6fa4: bioinformatics_pipeline_tutorial.lib.digest_protein_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/bXtyUfEfXnr080JI0000.fasta, hash=45bf7bec), enzyme_regex='[KR]', missed_cleavages=0, min_length=4, max_length=75) on default
[redun] Run Job 7f1e4017: bioinformatics_pipeline_tutorial.lib.digest_protein_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/QoNT2toucir3QJWw0000.fasta, hash=61d14102), enzyme_regex='[KR]', missed_cleavages=0, min_length=4, max_length=75) on default
[redun] Run Job 3b616f67: bioinformatics_pipeline_tutorial.lib.digest_protein_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/CtiEqWncr9uDvcbr0000.fasta, hash=ad3e6dc9), enzyme_regex='[KR]', missed_cleavages=0, min_length=4, max_length=75) on default
[redun] Run Job dc57375d: bioinformatics_pipeline_tutorial.lib.count_amino_acids_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/c44ljlhVZeSLf1W30000.fasta, hash=298f7f94), input_peptides=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/c44ljlhVZeSLf1W30000.peptides.txt, hash=1ff22443), amino_acid='C') on default
[redun] Run Job d6d3d100: bioinformatics_pipeline_tutorial.lib.count_amino_acids_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/bXtyUfEfXnr080JI0000.fasta, hash=45bf7bec), input_peptides=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/bXtyUfEfXnr080JI0000.peptides.txt, hash=c0bb1cf3), amino_acid='C') on default
[redun] Run Job 5dd35368: bioinformatics_pipeline_tutorial.lib.count_amino_acids_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/QoNT2toucir3QJWw0000.fasta, hash=61d14102), input_peptides=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/QoNT2toucir3QJWw0000.peptides.txt, hash=73fd1866), amino_acid='C') on default
[redun] Run Job e84b429e: bioinformatics_pipeline_tutorial.lib.count_amino_acids_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/CtiEqWncr9uDvcbr0000.fasta, hash=ad3e6dc9), input_peptides=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/CtiEqWncr9uDvcbr0000.peptides.txt, hash=cb036268), amino_acid='C') on default
[redun] Run Job 0f608433: bioinformatics_pipeline_tutorial.lib.plot_count_task(input_count=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/c44ljlhVZeSLf1W30000.count.tsv, hash=3b86cfe7)) on default
[redun] Run Job c92f3bca: bioinformatics_pipeline_tutorial.lib.plot_count_task(input_count=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/bXtyUfEfXnr080JI0000.count.tsv, hash=e91da858)) on default
[redun] Run Job 26ea85fe: bioinformatics_pipeline_tutorial.lib.plot_count_task(input_count=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/QoNT2toucir3QJWw0000.count.tsv, hash=96848f21)) on default
[redun] Run Job 65b1afab: bioinformatics_pipeline_tutorial.lib.plot_count_task(input_count=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/CtiEqWncr9uDvcbr0000.count.tsv, hash=24328a56)) on default
[redun] Run Job 1f2589ef: bioinformatics_pipeline_tutorial.lib.get_report_task(input_counts=[File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/c44ljlhVZeSLf1W30000.count.tsv, hash=3b86cfe7), File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-l...) on default
[redun] Run Job c09c9133: bioinformatics_pipeline_tutorial.lib.archive_results_task(inputs_plots=[File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/c44ljlhVZeSLf1W30000.plot.png, hash=43313f9f), File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-la..., input_report=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/protein_report.tsv, hash=660bcb4c)) on default
[redun] Run Job 9fb3a02b: redun_lamin_fasta.finish(results_archive=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/results.tgz, hash=4ad34654)) on default
[redun]
[redun] | JOB STATUS 2026/03/06 16:23:13
[redun] | TASK PENDING RUNNING FAILED CACHED DONE TOTAL
[redun] |
[redun] | ALL 0 0 0 0 16 16
[redun] | bioinformatics_pipeline_tutorial.lib.archive_results_task 0 0 0 0 1 1
[redun] | bioinformatics_pipeline_tutorial.lib.count_amino_acids_task 0 0 0 0 4 4
[redun] | bioinformatics_pipeline_tutorial.lib.digest_protein_task 0 0 0 0 4 4
[redun] | bioinformatics_pipeline_tutorial.lib.get_report_task 0 0 0 0 1 1
[redun] | bioinformatics_pipeline_tutorial.lib.plot_count_task 0 0 0 0 4 4
[redun] | redun_lamin_fasta.finish 0 0 0 0 1 1
[redun] | redun_lamin_fasta.main 0 0 0 0 1 1
[redun]
[redun]
[redun] Execution duration: 14.92 seconds
View data lineage¶
artifact = ln.Artifact.get(key="data/results.tgz")
artifact.view_lineage()
Show code cell output
artifact.transform.describe()
Show code cell output
Transform: workflow.py (0000) | description: CLI: redun ├── uid: 75jlbxVuCz0i0000 │ hash: zjZMEeofUMp3FV6fxbgjYg type: script │ branch: main space: all │ created_at: 2026-03-06 16:22:58 UTC created_by: testuser1 └── source_code: │ """workflow.py.""" │ │ # This code is based on a copy from https://github.com/ricomnl/bioinformatics-pi … │ # Copyright Rico Meinl 2022 │ from enum import Enum │ │ import lamindb as ln │ from redun import File, task │ │ import redun_lamin_fasta │ from redun_lamin_fasta.lib import ( │ archive_results_task, │ count_amino_acids_task, │ digest_protein_task, │ get_report_task, │ plot_count_task, │ ) │ │ redun_namespace = redun_lamin_fasta.__name__ │ │ │ class Executor(str, Enum): │ default = "default" │ process = "process" │ batch = "batch" │ batch_debug = "batch_debug" │ │ │ @task() │ def finish(results_archive: File) -> File: │ …
artifact.run.describe()
Show code cell output
Run: Bz2tiet (workflow.py) ├── uid: Bz2tietYvi0bBPtS transform: workflow.py (0000) │ | description: CLI: redun │ started_at: 2026-03-06 16:22:58 UTC finished_at: 2026-03-06 16:23:13 UTC │ status: completed │ branch: main space: all │ created_at: 2026-03-06 16:22:58 UTC created_by: testuser1 ├── cli_args: │ │ run workflow.py main --input-dir ./fasta --tag run=test-run ├── report: YzXLXyQ │ │ → connected lamindb: testuser1/test-redun-lamin │ │ → created Transform('75jlbxVuCz0i0000', key='workflow.py'), started new Run('Bz2 … │ │ • recommendation: to identify the script across renames, pass the uid: ln.track( … └── environment: XLMMGUq │ aiobotocore==2.26.0 │ aiohappyeyeballs==2.6.1 │ aiohttp==3.13.3 │ aioitertools==0.13.0 │ …
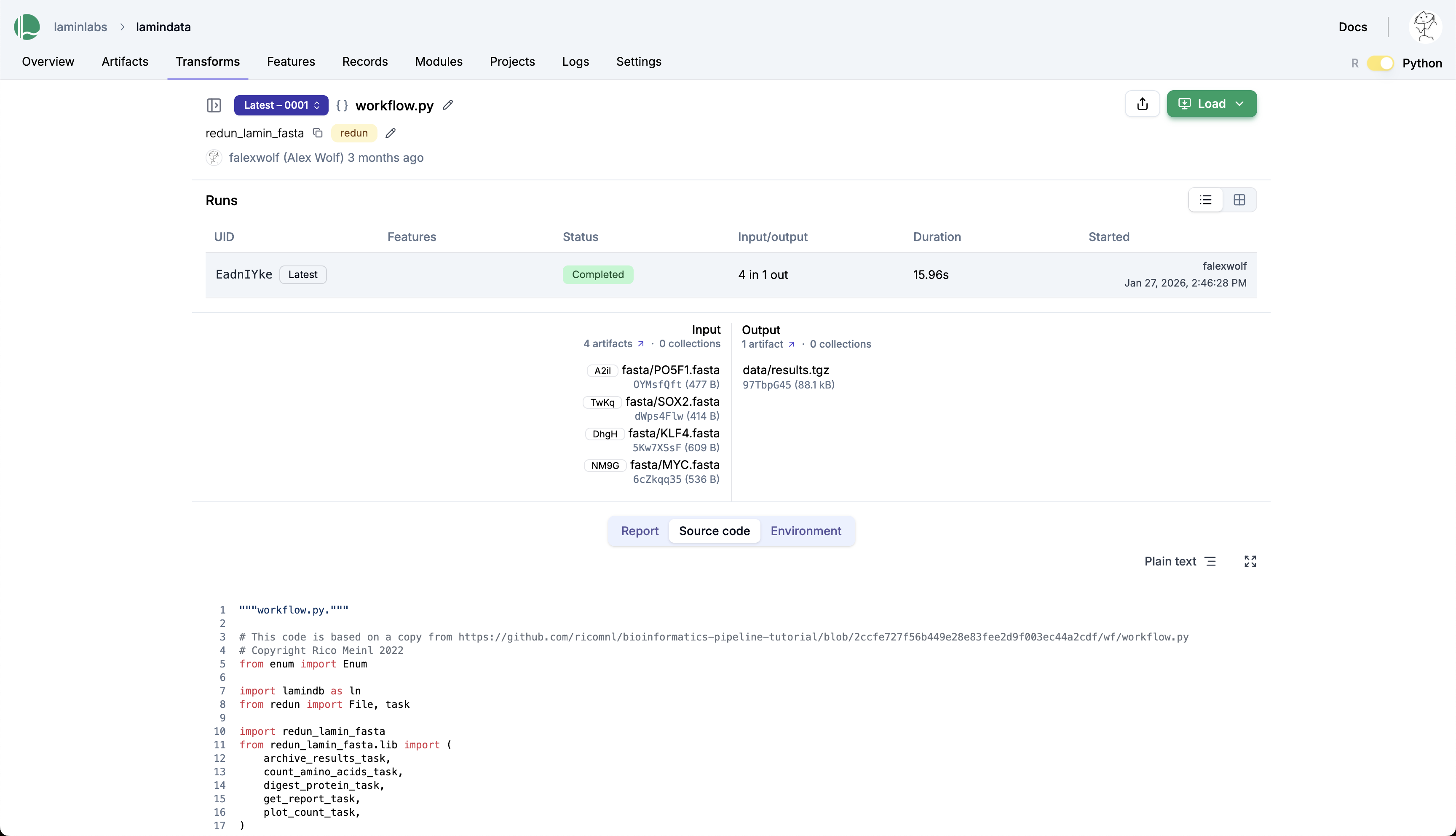
Explore the run on the hub¶
→ lamin.ai/laminlabs/lamindata/transform/taasWKawCiNA
View the database content¶
ln.view()
Show code cell output
Artifact
| uid | key | description | suffix | kind | otype | size | hash | n_files | n_observations | ... | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | storage_id | run_id | schema_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 6 | eLMBK7fw6ofSnerm0000 | data/results.tgz | None | .tgz | None | None | 94353 | EVKM7v96JtxMdIw7x-uA_Q | None | None | ... | True | False | 2026-03-06 16:23:13.319000+00:00 | 1 | 1 | 1 | 1 | 1.0 | None | 1 |
| 4 | CtiEqWncr9uDvcbr0000 | fasta/KLF4.fasta | None | .fasta | None | None | 609 | LyuoYkWs4SgYcH7P7JLJtA | None | None | ... | True | False | 2026-03-06 16:22:49.850000+00:00 | 1 | 1 | 1 | 1 | NaN | None | 1 |
| 3 | QoNT2toucir3QJWw0000 | fasta/PO5F1.fasta | None | .fasta | None | None | 477 | -7iJgveFO9ia0wE1bqVu6g | None | None | ... | True | False | 2026-03-06 16:22:49.850000+00:00 | 1 | 1 | 1 | 1 | NaN | None | 1 |
| 2 | bXtyUfEfXnr080JI0000 | fasta/MYC.fasta | None | .fasta | None | None | 536 | WGbEtzPw-3bQEGcngO_pHQ | None | None | ... | True | False | 2026-03-06 16:22:49.849000+00:00 | 1 | 1 | 1 | 1 | NaN | None | 1 |
| 1 | c44ljlhVZeSLf1W30000 | fasta/SOX2.fasta | None | .fasta | None | None | 414 | C5q_yaFXGk4SAEpfdqBwnQ | None | None | ... | True | False | 2026-03-06 16:22:49.848000+00:00 | 1 | 1 | 1 | 1 | NaN | None | 1 |
5 rows × 21 columns
Run
| uid | name | description | entrypoint | started_at | finished_at | params | reference | reference_type | cli_args | ... | created_at | branch_id | created_on_id | space_id | transform_id | report_id | environment_id | plan_id | created_by_id | initiated_by_run_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 1 | Bz2tietYvi0bBPtS | None | None | None | 2026-03-06 16:22:58.646153+00:00 | 2026-03-06 16:23:13.350451+00:00 | None | None | None | run workflow.py main --input-dir ./fasta --tag... | ... | 2026-03-06 16:22:58.647000+00:00 | 1 | 1 | 1 | 1 | 7 | 5 | None | 1 | None |
1 rows × 21 columns
Storage
| uid | root | description | type | region | instance_uid | is_locked | created_at | branch_id | created_on_id | space_id | created_by_id | run_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||
| 1 | mLNfMoHo6RPv | /home/runner/work/redun-lamin/redun-lamin/docs... | None | local | None | iQlBPgD8uaqR | False | 2026-03-06 16:22:46.418000+00:00 | 1 | 1 | 1 | 1 | None |
Transform
| uid | key | description | kind | source_code | hash | reference | reference_type | version_tag | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | environment_id | plan_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||||
| 1 | 75jlbxVuCz0i0000 | workflow.py | CLI: redun | script | """workflow.py."""\n\n# This code is based on ... | zjZMEeofUMp3FV6fxbgjYg | None | None | None | True | False | 2026-03-06 16:22:58.644000+00:00 | 1 | 1 | 1 | None | None | 1 |
Appendix¶
Map the redun execution id¶
If we want to be able to query LaminDB for redun execution ID, this here is a way to get it:
# export the run information from redun
!redun log --exec --exec-tag run=test-run --format json --no-pager > redun_exec.json
# load the redun execution id from the JSON and store it in the LaminDB run record
with open("redun_exec.json") as file:
redun_exec = json.loads(file.readline())
artifact.run.reference = redun_exec["id"]
artifact.run.reference_type = "redun_id"
artifact.run.save()
Show code cell output
Run(uid='Bz2tietYvi0bBPtS', name=None, description=None, entrypoint=None, started_at=2026-03-06 16:22:58 UTC, finished_at=2026-03-06 16:23:13 UTC, params=None, reference='6d368a45-6525-469c-8328-b76b622ebf64', reference_type='redun_id', cli_args='run workflow.py main --input-dir ./fasta --tag run=test-run', branch_id=1, created_on_id=1, space_id=1, transform_id=1, report_id=7, environment_id=5, plan_id=None, created_by_id=1, initiated_by_run_id=None, created_at=2026-03-06 16:22:58 UTC, is_locked=False)
Map the redun run report¶
While lamindb auto-tracks the logs of the main python process you might also want to link the dedicated redun logs:
report = ln.Artifact(
"run_logs.txt",
description=f"Redun run report of {redun_exec['id']}",
run=False,
kind="__lamindb_run__", # mark as auxiliary artifact for the run
).save()
artifact.run.report = report
artifact.run.save()
Show code cell output
Run(uid='Bz2tietYvi0bBPtS', name=None, description=None, entrypoint=None, started_at=2026-03-06 16:22:58 UTC, finished_at=2026-03-06 16:23:13 UTC, params=None, reference='6d368a45-6525-469c-8328-b76b622ebf64', reference_type='redun_id', cli_args='run workflow.py main --input-dir ./fasta --tag run=test-run', branch_id=1, created_on_id=1, space_id=1, transform_id=1, report_id=8, environment_id=5, plan_id=None, created_by_id=1, initiated_by_run_id=None, created_at=2026-03-06 16:22:58 UTC, is_locked=False)