RxRx: cell imaging 
 ¶
¶
rxrx.ai hosts high-throughput cell imaging datasets generated by Recursion.
High numbers of fluorescent microscopy images characterize cellular phenotypes in vitro based on morphology and protein expression (5-10 stains) across a range of conditions.
In this guide, you’ll see how to query some of these data using LaminDB. If you’d like to transfer data into your own LaminDB instance, see the transfer guide.
# !pip install 'lamindb[gcp]' duckdb
!lamin init --modules bionty,pertdb --storage ./test-rxrx
Show code cell output
→ initialized lamindb: testuser1/test-rxrx
import lamindb as ln
Show code cell output
→ connected lamindb: testuser1/test-rxrx
Create the central query object for this instance:
db = ln.DB("laminlabs/lamindata")
Search & look up metadata¶
We’ll find all genetic treatments in the GeneticPerturbation registry:
df = db.pertdb.GeneticPerturbation.to_dataframe()
df.shape
Show code cell output
(8, 17)
Let us create a look up object for siRNAs so that we can easily auto-complete queries involving it:
sirnas = db.pertdb.GeneticPerturbation.filter(type="siRNA").lookup(return_field="name")
We’re also interested in cell lines:
cell_lines = db.bionty.CellLine.lookup(return_field="abbr")
Load the collection¶
This is RxRx1: 125k images for 1138 siRNA perturbation across 4 cell lines reading out 5 stains, image dimension is 512x512x6.
Let us get the corresponding object and some information about it:
collection = db.Collection.get("Br2Z1lVSQBAkkbbt7ILu")
collection.view_lineage()
collection.describe()
Show code cell output
Collection: Annotated RxRx1 images (1) └── uid: Br2Z1lVSQBAkkbbt7ILu run: 2024-06-17T12:31:43.923373+00:00 (01-rxrx1-ingest.ipynb) branch: main space: all created_at: 2024-06-17 12:43:02 UTC created_by: sunnyosun
The dataset consists in a metadata file and a folder path pointing to the image files:
collection.meta_artifact.load().head()
Show code cell output
! run input wasn't tracked, call `ln.track()` and re-run
| site_id | well_id | cell_line | split | experiment | plate | well | site | well_type | sirna | sirna_id | path | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | HEPG2-08_1_B02_1 | HEPG2-08_1_B02 | HEPG2 | test | HEPG2-08 | 1 | B02 | 1 | negative_control | EMPTY | 1138 | images/test/HEPG2-08/Plate1/B02_s1_w1.png |
| 1 | HEPG2-08_1_B02_1 | HEPG2-08_1_B02 | HEPG2 | test | HEPG2-08 | 1 | B02 | 1 | negative_control | EMPTY | 1138 | images/test/HEPG2-08/Plate1/B02_s1_w2.png |
| 2 | HEPG2-08_1_B02_1 | HEPG2-08_1_B02 | HEPG2 | test | HEPG2-08 | 1 | B02 | 1 | negative_control | EMPTY | 1138 | images/test/HEPG2-08/Plate1/B02_s1_w3.png |
| 3 | HEPG2-08_1_B02_1 | HEPG2-08_1_B02 | HEPG2 | test | HEPG2-08 | 1 | B02 | 1 | negative_control | EMPTY | 1138 | images/test/HEPG2-08/Plate1/B02_s1_w4.png |
| 4 | HEPG2-08_1_B02_1 | HEPG2-08_1_B02 | HEPG2 | test | HEPG2-08 | 1 | B02 | 1 | negative_control | EMPTY | 1138 | images/test/HEPG2-08/Plate1/B02_s1_w5.png |
Query image files¶
Because we didn’t choose to register each image as a record in the Artifact registry, we have to query the images through the metadata file of the dataset:
df = collection.meta_artifact.load()
Show code cell output
! run input wasn't tracked, call `ln.track()` and re-run
We can query a subset of images using metadata registries & pandas query syntax:
query = df[(df.cell_line == cell_lines.hep_g2_cell) & (df.plate == 1) & (df.site == 2)]
query
Show code cell output
| site_id | well_id | cell_line | split | experiment | plate | well | site | well_type | sirna | sirna_id | path | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | HEPG2-08_1_B02_2 | HEPG2-08_1_B02 | HEPG2 | test | HEPG2-08 | 1 | B02 | 2 | negative_control | EMPTY | 1138 | images/test/HEPG2-08/Plate1/B02_s2_w1.png |
| 7 | HEPG2-08_1_B02_2 | HEPG2-08_1_B02 | HEPG2 | test | HEPG2-08 | 1 | B02 | 2 | negative_control | EMPTY | 1138 | images/test/HEPG2-08/Plate1/B02_s2_w2.png |
| 8 | HEPG2-08_1_B02_2 | HEPG2-08_1_B02 | HEPG2 | test | HEPG2-08 | 1 | B02 | 2 | negative_control | EMPTY | 1138 | images/test/HEPG2-08/Plate1/B02_s2_w3.png |
| 9 | HEPG2-08_1_B02_2 | HEPG2-08_1_B02 | HEPG2 | test | HEPG2-08 | 1 | B02 | 2 | negative_control | EMPTY | 1138 | images/test/HEPG2-08/Plate1/B02_s2_w4.png |
| 10 | HEPG2-08_1_B02_2 | HEPG2-08_1_B02 | HEPG2 | test | HEPG2-08 | 1 | B02 | 2 | negative_control | EMPTY | 1138 | images/test/HEPG2-08/Plate1/B02_s2_w5.png |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 358111 | HEPG2-07_1_O23_2 | HEPG2-07_1_O23 | HEPG2 | train | HEPG2-07 | 1 | O23 | 2 | treatment | s27411 | 489 | images/train/HEPG2-07/Plate1/O23_s2_w2.png |
| 358112 | HEPG2-07_1_O23_2 | HEPG2-07_1_O23 | HEPG2 | train | HEPG2-07 | 1 | O23 | 2 | treatment | s27411 | 489 | images/train/HEPG2-07/Plate1/O23_s2_w3.png |
| 358113 | HEPG2-07_1_O23_2 | HEPG2-07_1_O23 | HEPG2 | train | HEPG2-07 | 1 | O23 | 2 | treatment | s27411 | 489 | images/train/HEPG2-07/Plate1/O23_s2_w4.png |
| 358114 | HEPG2-07_1_O23_2 | HEPG2-07_1_O23 | HEPG2 | train | HEPG2-07 | 1 | O23 | 2 | treatment | s27411 | 489 | images/train/HEPG2-07/Plate1/O23_s2_w5.png |
| 358115 | HEPG2-07_1_O23_2 | HEPG2-07_1_O23 | HEPG2 | train | HEPG2-07 | 1 | O23 | 2 | treatment | s27411 | 489 | images/train/HEPG2-07/Plate1/O23_s2_w6.png |
20328 rows × 12 columns
To access the individual images based on this query result:
collection.data_artifact.storage.root
Show code cell output
'gs://rxrx1-europe-west4'
images = [f"{collection.data_artifact.storage.root}/{key}" for key in query.path]
images
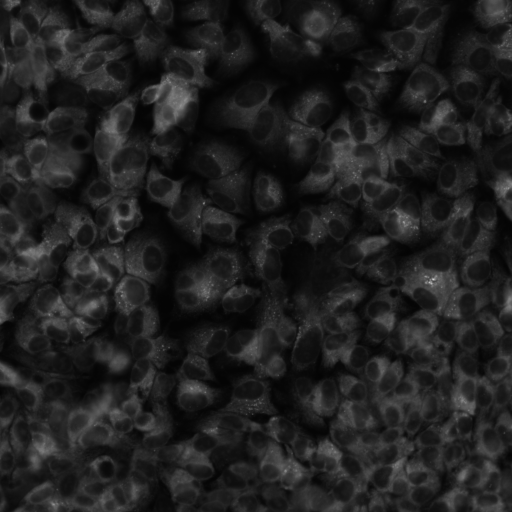
Download an image to disk:
from IPython.display import Image
path = ln.UPath(images[1])
path.download_to(".")
Image(f"./{path.name}")
Use DuckDB to query metadata¶
As an alternative to pandas, we could use DuckDB to query image metadata.
import duckdb
features = db.Feature.lookup(return_field="name")
filter = (
f"{features.cell_line} == '{cell_lines.hep_g2_cell}' and "
f"{features.plate} == '1' and {features.site} == '2'"
)
parquet_data = duckdb.from_parquet(
collection.meta_artifact.path.as_posix() + "?s3_region=us-east-1"
)
parquet_data.filter(filter)
Show code cell output
┌──────────────────┬────────────────┬───────────┬─────────┬────────────┬───────┬─────────┬───────┬──────────────────┬─────────┬──────────┬────────────────────────────────────────────┐
│ site_id │ well_id │ cell_line │ split │ experiment │ plate │ well │ site │ well_type │ sirna │ sirna_id │ path │
│ varchar │ varchar │ varchar │ varchar │ varchar │ int64 │ varchar │ int64 │ varchar │ varchar │ int64 │ varchar │
├──────────────────┼────────────────┼───────────┼─────────┼────────────┼───────┼─────────┼───────┼──────────────────┼─────────┼──────────┼────────────────────────────────────────────┤
│ HEPG2-08_1_B02_2 │ HEPG2-08_1_B02 │ HEPG2 │ test │ HEPG2-08 │ 1 │ B02 │ 2 │ negative_control │ EMPTY │ 1138 │ images/test/HEPG2-08/Plate1/B02_s2_w1.png │
│ HEPG2-08_1_B02_2 │ HEPG2-08_1_B02 │ HEPG2 │ test │ HEPG2-08 │ 1 │ B02 │ 2 │ negative_control │ EMPTY │ 1138 │ images/test/HEPG2-08/Plate1/B02_s2_w2.png │
│ HEPG2-08_1_B02_2 │ HEPG2-08_1_B02 │ HEPG2 │ test │ HEPG2-08 │ 1 │ B02 │ 2 │ negative_control │ EMPTY │ 1138 │ images/test/HEPG2-08/Plate1/B02_s2_w3.png │
│ HEPG2-08_1_B02_2 │ HEPG2-08_1_B02 │ HEPG2 │ test │ HEPG2-08 │ 1 │ B02 │ 2 │ negative_control │ EMPTY │ 1138 │ images/test/HEPG2-08/Plate1/B02_s2_w4.png │
│ HEPG2-08_1_B02_2 │ HEPG2-08_1_B02 │ HEPG2 │ test │ HEPG2-08 │ 1 │ B02 │ 2 │ negative_control │ EMPTY │ 1138 │ images/test/HEPG2-08/Plate1/B02_s2_w5.png │
│ HEPG2-08_1_B02_2 │ HEPG2-08_1_B02 │ HEPG2 │ test │ HEPG2-08 │ 1 │ B02 │ 2 │ negative_control │ EMPTY │ 1138 │ images/test/HEPG2-08/Plate1/B02_s2_w6.png │
│ HEPG2-08_1_B03_2 │ HEPG2-08_1_B03 │ HEPG2 │ test │ HEPG2-08 │ 1 │ B03 │ 2 │ treatment │ s21721 │ 855 │ images/test/HEPG2-08/Plate1/B03_s2_w1.png │
│ HEPG2-08_1_B03_2 │ HEPG2-08_1_B03 │ HEPG2 │ test │ HEPG2-08 │ 1 │ B03 │ 2 │ treatment │ s21721 │ 855 │ images/test/HEPG2-08/Plate1/B03_s2_w2.png │
│ HEPG2-08_1_B03_2 │ HEPG2-08_1_B03 │ HEPG2 │ test │ HEPG2-08 │ 1 │ B03 │ 2 │ treatment │ s21721 │ 855 │ images/test/HEPG2-08/Plate1/B03_s2_w3.png │
│ HEPG2-08_1_B03_2 │ HEPG2-08_1_B03 │ HEPG2 │ test │ HEPG2-08 │ 1 │ B03 │ 2 │ treatment │ s21721 │ 855 │ images/test/HEPG2-08/Plate1/B03_s2_w4.png │
│ · │ · │ · │ · │ · │ · │ · │ · │ · │ · │ · │ · │
│ · │ · │ · │ · │ · │ · │ · │ · │ · │ · │ · │ · │
│ · │ · │ · │ · │ · │ · │ · │ · │ · │ · │ · │ · │
│ HEPG2-02_1_G17_2 │ HEPG2-02_1_G17 │ HEPG2 │ train │ HEPG2-02 │ 1 │ G17 │ 2 │ treatment │ s27876 │ 3 │ images/train/HEPG2-02/Plate1/G17_s2_w1.png │
│ HEPG2-02_1_G17_2 │ HEPG2-02_1_G17 │ HEPG2 │ train │ HEPG2-02 │ 1 │ G17 │ 2 │ treatment │ s27876 │ 3 │ images/train/HEPG2-02/Plate1/G17_s2_w2.png │
│ HEPG2-02_1_G17_2 │ HEPG2-02_1_G17 │ HEPG2 │ train │ HEPG2-02 │ 1 │ G17 │ 2 │ treatment │ s27876 │ 3 │ images/train/HEPG2-02/Plate1/G17_s2_w3.png │
│ HEPG2-02_1_G17_2 │ HEPG2-02_1_G17 │ HEPG2 │ train │ HEPG2-02 │ 1 │ G17 │ 2 │ treatment │ s27876 │ 3 │ images/train/HEPG2-02/Plate1/G17_s2_w4.png │
│ HEPG2-02_1_G17_2 │ HEPG2-02_1_G17 │ HEPG2 │ train │ HEPG2-02 │ 1 │ G17 │ 2 │ treatment │ s27876 │ 3 │ images/train/HEPG2-02/Plate1/G17_s2_w5.png │
│ HEPG2-02_1_G17_2 │ HEPG2-02_1_G17 │ HEPG2 │ train │ HEPG2-02 │ 1 │ G17 │ 2 │ treatment │ s27876 │ 3 │ images/train/HEPG2-02/Plate1/G17_s2_w6.png │
│ HEPG2-02_1_G18_2 │ HEPG2-02_1_G18 │ HEPG2 │ train │ HEPG2-02 │ 1 │ G18 │ 2 │ treatment │ s27646 │ 124 │ images/train/HEPG2-02/Plate1/G18_s2_w1.png │
│ HEPG2-02_1_G18_2 │ HEPG2-02_1_G18 │ HEPG2 │ train │ HEPG2-02 │ 1 │ G18 │ 2 │ treatment │ s27646 │ 124 │ images/train/HEPG2-02/Plate1/G18_s2_w2.png │
│ HEPG2-02_1_G18_2 │ HEPG2-02_1_G18 │ HEPG2 │ train │ HEPG2-02 │ 1 │ G18 │ 2 │ treatment │ s27646 │ 124 │ images/train/HEPG2-02/Plate1/G18_s2_w3.png │
│ HEPG2-02_1_G18_2 │ HEPG2-02_1_G18 │ HEPG2 │ train │ HEPG2-02 │ 1 │ G18 │ 2 │ treatment │ s27646 │ 124 │ images/train/HEPG2-02/Plate1/G18_s2_w4.png │
├──────────────────┴────────────────┴───────────┴─────────┴────────────┴───────┴─────────┴───────┴──────────────────┴─────────┴──────────┴────────────────────────────────────────────┤
│ ? rows (>9999 rows, 20 shown) 12 columns │
└─────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────┘