Analyze the collection and save a result 
 ¶
¶
import lamindb as ln
import bionty as bt
ln.track("zzJzdgJ763Dy0000")
Show code cell output
→ connected lamindb: testuser1/test-facs
→ created Transform('zzJzdgJ763Dy0000', key='facs4.ipynb'), started new Run('mKVMb6udzHYF3Soq') at 2026-02-24 14:53:54 UTC
→ notebook imports: bionty==2.2.1 lamindb==2.2.1 scanpy==1.12
ln.Collection.to_dataframe()
Show code cell output
| uid | key | description | hash | reference | reference_type | version_tag | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | created_by_id | run_id | meta_artifact_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||
| 2 | zToIArXEOSKiwEif0001 | My versioned cytometry collection | None | eNSQbRNsjg278G1RwXmqCw | None | None | 2 | True | False | 2026-02-24 14:53:48.507000+00:00 | 1 | 1 | 1 | 1 | 2 | None |
| 1 | zToIArXEOSKiwEif0000 | My versioned cytometry collection | None | jZ8YvUdZGRwPgfoBQbOg_Q | None | None | 1 | False | False | 2026-02-24 14:53:39.038000+00:00 | 1 | 1 | 1 | 1 | 1 | None |
collection = ln.Collection.get(key="My versioned cytometry collection", version="2")
adata = collection.load(join="inner")
Show code cell output
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/anndata/_core/anndata.py:1823: UserWarning: Observation names are not unique. To make them unique, call `.obs_names_make_unique`.
utils.warn_names_duplicates("obs")
The AnnData has the reference to the individual files in the .obs annotations:
adata.obs.artifact_uid.cat.categories
Show code cell output
Index(['QDBjHCqE0E4oCdtf0000', 'ww96sdC4PfernY5y0000'], dtype='object')
By default, the intersection of features is used:
adata.var.index
Show code cell output
Index(['CD8', 'CD27', 'Ccr7', 'Cd4', 'CD45RA', 'CD3'], dtype='object')
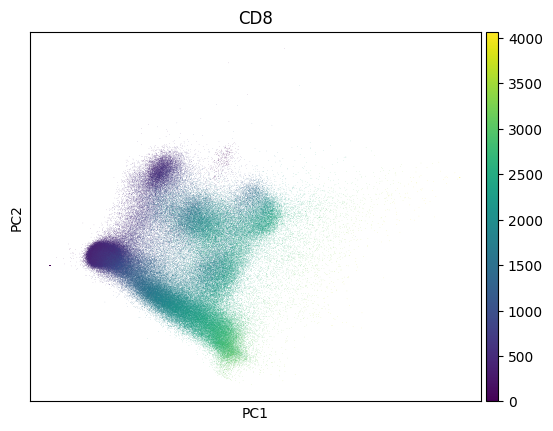
Let us create a plot:
markers = bt.CellMarker.lookup()
import scanpy as sc
sc.pp.pca(adata)
sc.pl.pca(adata, color=markers.cd8.name, save="_cd8")
Show code cell output
WARNING: saving figure to file figures/pca_cd8.pdf
/tmp/ipykernel_3652/1861229803.py:4: FutureWarning: Argument `save` is deprecated and will be removed in a future version. Use `sc.pl.plot(show=False).figure.savefig()` instead.
sc.pl.pca(adata, color=markers.cd8.name, save="_cd8")
artifact = ln.Artifact("./figures/pca_cd8.pdf", description="My result on CD8").save()
artifact.view_lineage()
Show code cell output
Given the image is part of the notebook, there isn’t an actual need to save it and you can also rely on the report that you’ll create when saving the notebook:
ln.finish()
# clean up test instance
!rm -r test-flow
!lamin delete --force test-facs
Show code cell output
rm: cannot remove 'test-flow': No such file or directory
╭─ Error ──────────────────────────────────────────────────────────────────────╮
│ '/home/runner/work/lamin-usecases/lamin-usecases/docs/test-facs/.lamindb' │
│ contains 3 objects: │
│ /home/runner/work/lamin-usecases/lamin-usecases/docs/test-facs/.lamindb/IHNj │
│ Az8zoLBfap7B0000.pdf │
│ /home/runner/work/lamin-usecases/lamin-usecases/docs/test-facs/.lamindb/QDBj │
│ HCqE0E4oCdtf0000.h5ad │
│ /home/runner/work/lamin-usecases/lamin-usecases/docs/test-facs/.lamindb/ww96 │
│ sdC4PfernY5y0000.h5ad │
│ delete them prior to deleting the storage location │
╰──────────────────────────────────────────────────────────────────────────────╯