Nextflow 
 ¶
¶
Nextflow is the most widely used workflow manager in bioinformatics.
This guide shows how to register a nf-core/scrnaseq Nextflow run using the nf-lamin plugin.
See Post-run script learn how to register a Nextflow run using a post-run script.
To use the nf-lamin plugin, you need to configure it with your LaminDB instance and API key.
This setup allows the plugin to authenticate and interact with your LaminDB instance, enabling it to record workflow runs and associated metadata.
Set API Key¶
Retrieve your Lamin API key from your Lamin Hub account settings and set it as a Nextflow secret:
nextflow secrets set LAMIN_API_KEY <your-lamin-api-key>
Configure the plugin¶
Add the following block to your nextflow.config:
plugins {
id 'nf-lamin'
}
lamin {
instance = "<your-lamin-org>/<your-lamin-instance>"
api_key = secrets.LAMIN_API_KEY
}
See nf-lamin plugin reference for more configuration options.
Example Run with nf-core/scrnaseq¶
This guide shows how to register a Nextflow run with inputs & outputs for the nf-core/scrnaseq pipeline.
Run the pipeline¶
With the nf-lamin plugin configured, let’s run the nf-core/scrnaseq pipeline on remote input data.
# The test profile uses publicly available test data
!nextflow run nf-core/scrnaseq \
-r "4.0.0" \
-profile docker,test \
-plugins nf-lamin \
--outdir s3://lamindb-ci/nf-lamin/run_$(date +%Y%m%d_%H%M%S)
Show code cell output
N E X T F L O W ~ version 25.10.4
Pulling nf-core/scrnaseq ...
downloaded from https://github.com/nf-core/scrnaseq.git
Downloading plugin [email protected]
Launching `https://github.com/nf-core/scrnaseq` [trusting_euclid] DSL2 - revision: e0ddddbff9 [4.0.0]
Downloading plugin [email protected]
Downloading plugin [email protected]
------------------------------------------------------
,--./,-.
___ __ __ __ ___ /,-._.--~'
|\ | |__ __ / ` / \ |__) |__ } {
| \| | \__, \__/ | \ |___ \`-._,-`-,
`._,._,'
nf-core/scrnaseq 4.0.0
------------------------------------------------------
Input/output options
input : https://github.com/nf-core/test-datasets/raw/scrnaseq/samplesheet-2-0.csv
outdir : s3://lamindb-ci/nf-lamin/run_20260303_184310
Mandatory arguments
aligner : star
protocol : 10XV2
Skip Tools
skip_cellbender : true
Reference genome options
fasta : https://github.com/nf-core/test-datasets/raw/scrnaseq/reference/GRCm38.p6.genome.chr19.fa
gtf : https://github.com/nf-core/test-datasets/raw/scrnaseq/reference/gencode.vM19.annotation.chr19.gtf
save_align_intermeds : true
Institutional config options
config_profile_name : Test profile
config_profile_description: Minimal test dataset to check pipeline function
Generic options
trace_report_suffix : 2026-03-03_18-43-22
Core Nextflow options
revision : 4.0.0
runName : trusting_euclid
containerEngine : docker
launchDir : /home/runner/work/nf-lamin/nf-lamin/docs
workDir : /home/runner/work/nf-lamin/nf-lamin/docs/work
projectDir : /home/runner/.nextflow/assets/nf-core/scrnaseq
userName : runner
profile : docker,test
configFiles : /home/runner/.nextflow/assets/nf-core/scrnaseq/nextflow.config, /home/runner/work/nf-lamin/nf-lamin/docs/nextflow.config
!! Only displaying parameters that differ from the pipeline defaults !!
------------------------------------------------------
* The pipeline
https://doi.org/10.5281/zenodo.3568187
* The nf-core framework
https://doi.org/10.1038/s41587-020-0439-x
* Software dependencies
https://github.com/nf-core/scrnaseq/blob/master/CITATIONS.md
WARN: The following invalid input values have been detected:
* --validationSchemaIgnoreParams: genomes
[a2/762a93] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:FASTQC_CHECK:FASTQC (Sample_X)
[b1/cdb2cf] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:FASTQC_CHECK:FASTQC (Sample_Y)
[a3/50b23d] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:GTF_GENE_FILTER (GRCm38.p6.genome.chr19.fa)
[b5/3e1291] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:STARSOLO:STAR_GENOMEGENERATE (GRCm38.p6.genome.chr19.fa)
[1e/9cc135] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:STARSOLO:STAR_ALIGN (Sample_X)
[9d/d72b84] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:STARSOLO:STAR_ALIGN (Sample_Y)
[14/fe7d02] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:MTX_TO_H5AD (Sample_X)
[b6/dd7d28] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:MTX_TO_H5AD (Sample_X)
[73/8bd7ee] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:MTX_TO_H5AD (Sample_Y)
[e6/9caa98] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:MTX_TO_H5AD (Sample_Y)
[ec/377aa2] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:H5AD_CONVERSION:ANNDATAR_CONVERT (Sample_X)
WARN: Unable to stage foreign file: https://github.com/nf-core/test-datasets/raw/scrnaseq/samplesheet-2-0.csv (try 1 of 3) -- Cause: Unable to access path: https://github.com/nf-core/test-datasets/raw/scrnaseq/samplesheet-2-0.csv
WARN: Unable to stage foreign file: https://github.com/nf-core/test-datasets/raw/scrnaseq/samplesheet-2-0.csv (try 2 of 3) -- Cause: Unable to access path: https://github.com/nf-core/test-datasets/raw/scrnaseq/samplesheet-2-0.csv
WARN: Unable to stage foreign file: https://github.com/nf-core/test-datasets/raw/scrnaseq/samplesheet-2-0.csv (try 3 of 3) -- Cause: Unable to access path: https://github.com/nf-core/test-datasets/raw/scrnaseq/samplesheet-2-0.csv
ERROR ~ Error executing process > 'NFCORE_SCRNASEQ:SCRNASEQ:H5AD_CONVERSION:CONCAT_H5AD (1)'
Caused by:
Can't stage file https://github.com/nf-core/test-datasets/raw/scrnaseq/samplesheet-2-0.csv -- reason: Unable to access path: https://github.com/nf-core/test-datasets/raw/scrnaseq/samplesheet-2-0.csv
Command executed [/home/runner/.nextflow/assets/nf-core/scrnaseq/modules/local/templates/concat_h5ad.py]:
#!/usr/bin/env python
# Set numba chache dir to current working directory (which is a writable mount also in containers)
import os
os.environ["NUMBA_CACHE_DIR"] = "."
import scanpy as sc, anndata as ad, pandas as pd
from pathlib import Path
import platform
def read_samplesheet(samplesheet):
df = pd.read_csv(samplesheet)
df.set_index("sample")
# samplesheet may contain replicates, when it has,
# group information from replicates and collapse with commas
# only keep unique values using set()
df = df.groupby(["sample"]).agg(lambda column: ",".join(set(column.astype(str))))
return df
def format_yaml_like(data: dict, indent: int = 0) -> str:
"""Formats a dictionary to a YAML-like string.
Args:
data (dict): The dictionary to format.
indent (int): The current indentation level.
Returns:
str: A string formatted as YAML.
"""
yaml_str = ""
for key, value in data.items():
spaces = " " * indent
if isinstance(value, dict):
yaml_str += f"{spaces}{key}:\n{format_yaml_like(value, indent + 1)}"
else:
yaml_str += f"{spaces}{key}: {value}\n"
return yaml_str
def dump_versions():
versions = {
"NFCORE_SCRNASEQ:SCRNASEQ:H5AD_CONVERSION:CONCAT_H5AD": {
"python": platform.python_version(),
"scanpy": sc.__version__,
}
}
with open("versions.yml", "w") as f:
f.write(format_yaml_like(versions))
if __name__ == "__main__":
# Open samplesheet as dataframe
df_samplesheet = read_samplesheet("samplesheet-2-0.csv")
# find all h5ad and append to dict
dict_of_h5ad = {str(path).replace("_matrix.h5ad", ""): sc.read_h5ad(path) for path in Path(".").rglob("*.h5ad")}
# concat h5ad files
adata = ad.concat(dict_of_h5ad, label="sample", merge="unique", index_unique="_")
# merge with data.frame, on sample information
adata.obs = adata.obs.join(df_samplesheet, on="sample", how="left").astype(str)
adata.write_h5ad("combined_filtered_matrix.h5ad")
print("Wrote h5ad file to {}".format("combined_filtered_matrix.h5ad"))
# dump versions
dump_versions()
Command exit status:
-
Command output:
(empty)
Container:
community.wave.seqera.io/library/scanpy:1.10.2--e83da2205b92a538
Tip: you can try to figure out what's wrong by changing to the process work dir and showing the script file named `.command.sh`
-- Check '.nextflow.log' file for details
Execution cancelled -- Finishing pending tasks before exit
ERROR ~ Pipeline failed. Please refer to troubleshooting docs: https://nf-co.re/docs/usage/troubleshooting
-- Check '.nextflow.log' file for details
-[nf-core/scrnaseq] Pipeline completed with errors-
What is the full command and output when running this command?
nextflow run nf-core/scrnaseq \
-r "4.0.0" \
-profile docker \
-plugins nf-lamin \
--input https://github.com/nf-core/test-datasets/raw/scrnaseq/samplesheet-2-0.csv \
--fasta https://github.com/nf-core/test-datasets/raw/scrnaseq/reference/GRCm38.p6.genome.chr19.fa \
--gtf https://github.com/nf-core/test-datasets/raw/scrnaseq/reference/gencode.vM19.annotation.chr19.gtf \
--protocol 10XV2 \
--aligner star \
--skip_cellbender \
--outdir s3://lamindb-ci/nf-lamin/run_$(date +%Y%m%d_%H%M%S)
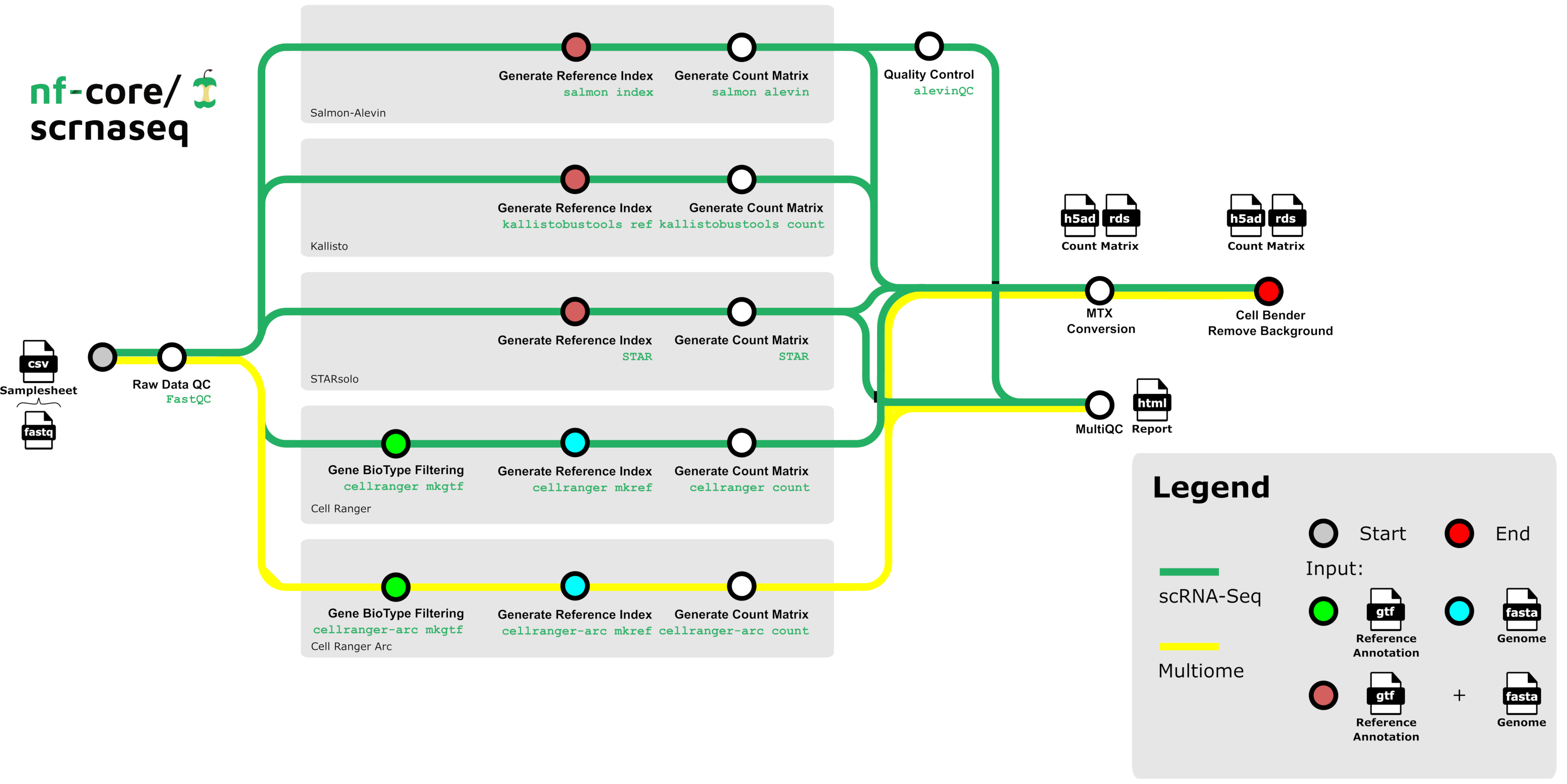
What steps are executed by the nf-core/scrnaseq pipeline?
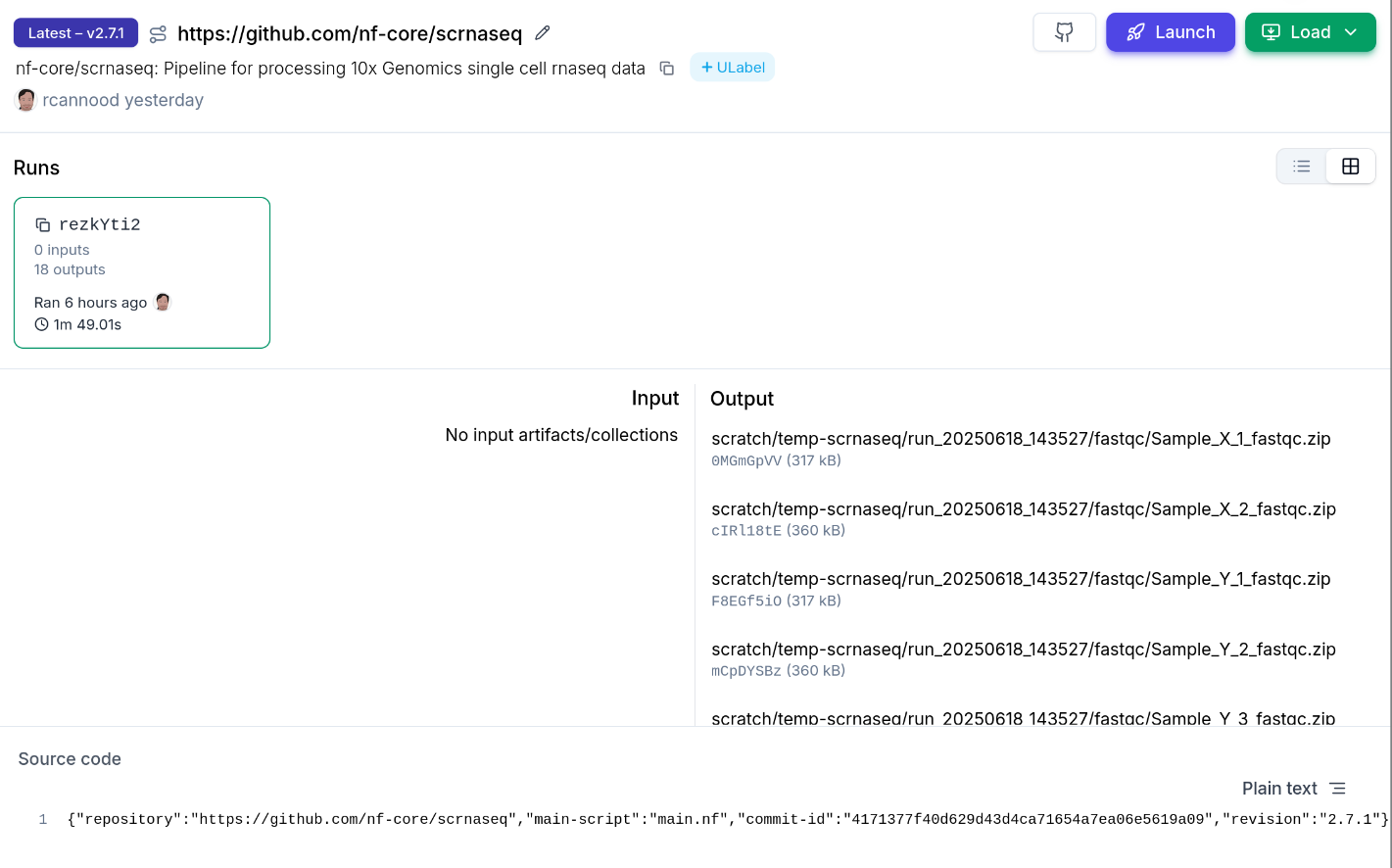
When you run this command, nf-lamin will print links to the Transform and Run records it creates in Lamin Hub:
✅ Connected to LaminDB instance 'laminlabs/lamindata' as 'user_name'
Transform J49HdErpEFrs0000 (https://staging.laminhub.com/laminlabs/lamindata/transform/J49HdErpEFrs0000)
Run p8npJ8JxIYazW4EkIl8d (https://staging.laminhub.com/laminlabs/lamindata/transform/J49HdErpEFrs0000/p8npJ8JxIYazW4EkIl8d)
View transforms & runs on Lamin Hub¶
You can explore the run and its associated artifacts through Lamin Hub or the Python package.
Via Lamin Hub¶
Transform: J49HdErpEFrs0000
Run: p8npJ8JxIYazW4EkIl8d

Using LaminDB¶
import lamindb as ln
# Make sure you are connected to the same instance
# you configured in nextflow.config
ln.Run.get("p8npJ8JxIYazW4EkIl8d")
This will display the details of the run record in your notebook:
Run(uid='p8npJ8JxIYazW4EkIl8d', name='trusting_brazil', started_at=2025-06-18 12:35:30 UTC, finished_at=2025-06-18 12:37:19 UTC, transform_id='aBcDeFg', created_by_id=..., created_at=...)