Re-constructing Schmidt et al. (2022)  ¶
¶
This use-case re-constructs a result of the research project of Schmidt el al. (2022) via several scripts & notebooks.
A genome-wide phenotypic CRISPRa screen was used to find transcriptional cell states that correlate with causal drivers of proteomic IFNG expression.
To understand how this result was obtained, one can look at the lineage:
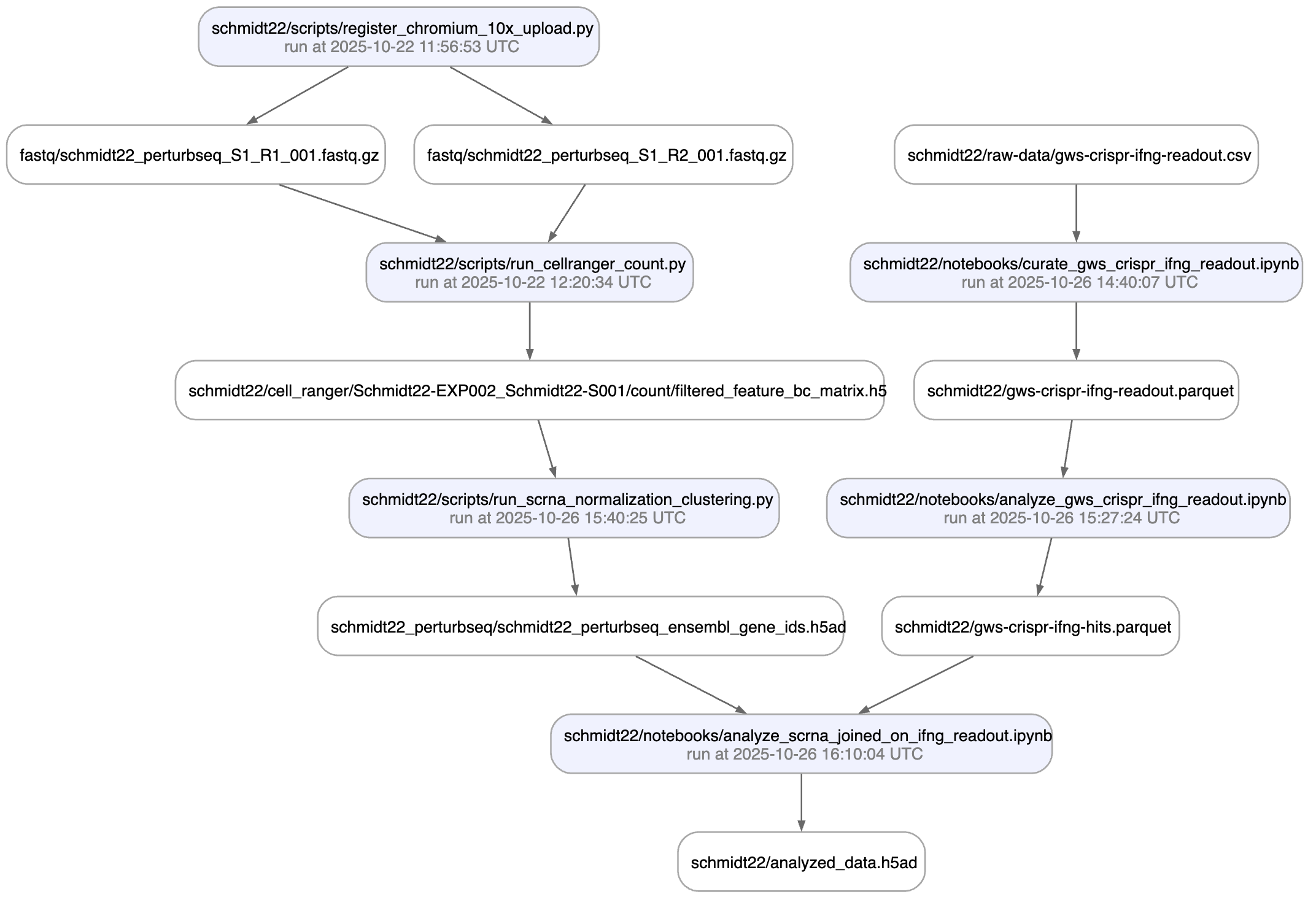
You can explore it here together with code and data artifacts.
You can also see all code and how it can be executed in build.yml here: