Post-run script 
 ¶
¶
Nextflow is the most widely used workflow manager in bioinformatics.
We generally recommend using the nf-lamin plugin.
However, if lower level LaminDB usage is required, it might be worthwhile writing a custom Python script.
This guide shows how to register a Nextflow run with inputs & outputs for the example of the nf-core/scrnaseq pipeline by running a Python script.
The approach could be automated by deploying the script via
a serverless environment trigger (e.g., AWS Lambda)
a post-run script on the Seqera Platform
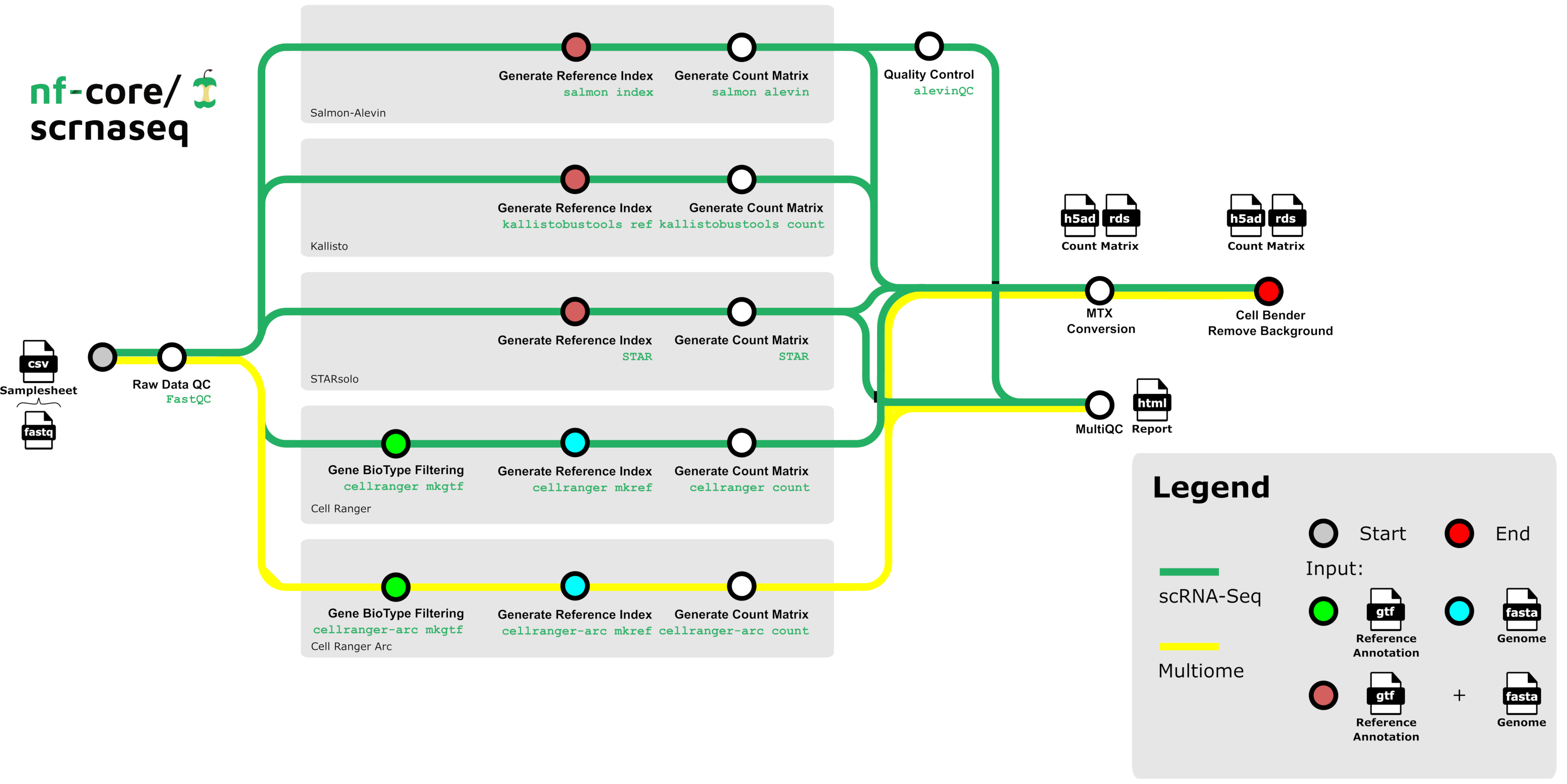
What steps are executed by the nf-core/scrnaseq pipeline?
!lamin init --storage ./test-nextflow --name test-nextflow
Show code cell output
→ initialized lamindb: testuser1/test-nextflow
Run the pipeline¶
Let’s download the input data from an S3 bucket.
import lamindb as ln
input_path = ln.UPath("s3://lamindb-test/scrnaseq_input")
input_path.download_to("scrnaseq_input")
Show code cell output
→ connected lamindb: testuser1/test-nextflow
And run the nf-core/scrnaseq pipeline.
# the test profile uses all downloaded input files as an input
!nextflow run nf-core/scrnaseq -r 4.0.0 -profile docker,test -resume --outdir scrnaseq_output
Show code cell output
N E X T F L O W ~ version 25.10.4
Pulling nf-core/scrnaseq ...
downloaded from https://github.com/nf-core/scrnaseq.git
WARN: It appears you have never run this project before -- Option `-resume` is ignored
Downloading plugin [email protected]
WARN: It appears you have never run this project before -- Option `-resume` is ignored
Launching `https://github.com/nf-core/scrnaseq` [wise_ritchie] DSL2 - revision: e0ddddbff9 [4.0.0]
Downloading plugin [email protected]
------------------------------------------------------
,--./,-.
___ __ __ __ ___ /,-._.--~'
|\ | |__ __ / ` / \ |__) |__ } {
| \| | \__, \__/ | \ |___ \`-._,-`-,
`._,._,'
nf-core/scrnaseq 4.0.0
------------------------------------------------------
Input/output options
input : https://github.com/nf-core/test-datasets/raw/scrnaseq/samplesheet-2-0.csv
outdir : scrnaseq_output
Mandatory arguments
aligner : star
protocol : 10XV2
Skip Tools
skip_cellbender : true
Reference genome options
fasta : https://github.com/nf-core/test-datasets/raw/scrnaseq/reference/GRCm38.p6.genome.chr19.fa
gtf : https://github.com/nf-core/test-datasets/raw/scrnaseq/reference/gencode.vM19.annotation.chr19.gtf
save_align_intermeds : true
Institutional config options
config_profile_name : Test profile
config_profile_description: Minimal test dataset to check pipeline function
Generic options
trace_report_suffix : 2026-02-19_11-05-13
Core Nextflow options
revision : 4.0.0
runName : wise_ritchie
containerEngine : docker
launchDir : /home/runner/work/nf-lamin/nf-lamin/docs
workDir : /home/runner/work/nf-lamin/nf-lamin/docs/work
projectDir : /home/runner/.nextflow/assets/nf-core/scrnaseq
userName : runner
profile : docker,test
configFiles : /home/runner/.nextflow/assets/nf-core/scrnaseq/nextflow.config, /home/runner/work/nf-lamin/nf-lamin/docs/nextflow.config
!! Only displaying parameters that differ from the pipeline defaults !!
------------------------------------------------------
* The pipeline
https://doi.org/10.5281/zenodo.3568187
* The nf-core framework
https://doi.org/10.1038/s41587-020-0439-x
* Software dependencies
https://github.com/nf-core/scrnaseq/blob/master/CITATIONS.md
Downloading plugin [email protected]
WARN: The following invalid input values have been detected:
* --validationSchemaIgnoreParams: genomes
[a0/f62127] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:FASTQC_CHECK:FASTQC (Sample_X)
[c8/ed2dbe] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:FASTQC_CHECK:FASTQC (Sample_Y)
[3b/5097d7] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:GTF_GENE_FILTER (GRCm38.p6.genome.chr19.fa)
[b7/6dfb84] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:STARSOLO:STAR_GENOMEGENERATE (GRCm38.p6.genome.chr19.fa)
[4b/fbea35] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:STARSOLO:STAR_ALIGN (Sample_X)
[5c/1d7922] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:STARSOLO:STAR_ALIGN (Sample_Y)
[0a/a9f6fe] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:MTX_TO_H5AD (Sample_X)
[8e/61329e] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:MTX_TO_H5AD (Sample_Y)
[da/2eb836] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:MTX_TO_H5AD (Sample_X)
[12/5dc489] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:MTX_TO_H5AD (Sample_Y)
[59/560b56] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:H5AD_CONVERSION:ANNDATAR_CONVERT (Sample_X)
[05/dc8e8c] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:MULTIQC
[a4/b1be96] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:H5AD_CONVERSION:ANNDATAR_CONVERT (Sample_Y)
[6d/6e8287] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:H5AD_CONVERSION:ANNDATAR_CONVERT (Sample_X)
[fd/5f05f3] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:H5AD_CONVERSION:ANNDATAR_CONVERT (Sample_Y)
[8a/db0e15] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:H5AD_CONVERSION:CONCAT_H5AD (combined)
[57/de552a] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:H5AD_CONVERSION:CONCAT_H5AD (combined)
[28/607ca7] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:H5AD_CONVERSION:ANNDATAR_CONVERT (combined)
[c7/f3d9ad] Submitted process > NFCORE_SCRNASEQ:SCRNASEQ:H5AD_CONVERSION:ANNDATAR_CONVERT (combined)
-[nf-core/scrnaseq] Pipeline completed successfully-
What is the full run command for the test profile?
nextflow run nf-core/scrnaseq -r 4.0.0 \
-profile docker \
-resume \
--outdir scrnaseq_output \
--input 'scrnaseq_input/samplesheet-2-0.csv' \
--skip_emptydrops \
--fasta 'https://github.com/nf-core/test-datasets/raw/scrnaseq/reference/GRCm38.p6.genome.chr19.fa' \
--gtf 'https://github.com/nf-core/test-datasets/raw/scrnaseq/reference/gencode.vM19.annotation.chr19.gtf' \
--aligner 'star' \
--protocol '10XV2' \
--max_cpus 2 \
--max_memory '6.GB' \
--max_time '6.h'
Run the registration script¶
After the pipeline has completed, a Python script registers inputs & outputs in LaminDB.
import argparse
import lamindb as ln
import json
import re
from pathlib import Path
from lamin_utils import logger
def parse_arguments() -> argparse.Namespace:
parser = argparse.ArgumentParser()
parser.add_argument("--input", type=str, required=True)
parser.add_argument("--output", type=str, required=True)
return parser.parse_args()
def register_pipeline_io(input_dir: str, output_dir: str, run: ln.Run) -> None:
"""Register input and output artifacts for an `nf-core/scrnaseq` run."""
input_artifacts = ln.Artifact.from_dir(input_dir, run=False)
ln.save(input_artifacts)
run.input_artifacts.set(input_artifacts)
ln.Artifact(f"{output_dir}/multiqc", description="multiqc report", run=run).save()
ln.Artifact(
f"{output_dir}/star/mtx_conversions/combined_filtered_matrix.h5ad",
key="filtered_count_matrix.h5ad",
run=run,
).save()
def register_pipeline_metadata(output_dir: str, run: ln.Run) -> None:
"""Register nf-core run metadata stored in the 'pipeline_info' folder."""
ulabel = ln.ULabel(name="nextflow").save()
run.transform.ulabels.add(ulabel)
# nextflow run id
content = next(Path(f"{output_dir}/pipeline_info").glob("execution_report_*.html")).read_text()
match = re.search(r"run id \[([^\]]+)\]", content)
nextflow_id = match.group(1) if match else ""
run.reference = nextflow_id
run.reference_type = "nextflow_id"
# completed at
completion_match = re.search(r'<span id="workflow_complete">([^<]+)</span>', content)
if completion_match:
from datetime import datetime
timestamp_str = completion_match.group(1).strip()
run.finished_at = datetime.strptime(timestamp_str, "%d-%b-%Y %H:%M:%S")
# execution report and software versions
for file_pattern, description, run_attr in [
("execution_report*", "execution report", "report"),
("nf_core_*_software*", "software versions", "environment"),
]:
matching_files = list(Path(f"{output_dir}/pipeline_info").glob(file_pattern))
if not matching_files:
logger.warning(f"No files matching '{file_pattern}' in pipeline_info")
continue
artifact = ln.Artifact(
matching_files[0],
description=f"nextflow run {description} of {nextflow_id}",
visibility=0,
run=False,
).save()
setattr(run, run_attr, artifact)
# nextflow run parameters
params_path = next(Path(f"{output_dir}/pipeline_info").glob("params*"))
with params_path.open() as params_file:
params = json.load(params_file)
ln.Param(name="params", dtype="dict").save()
run.features.add_values({"params": params})
run.save()
args = parse_arguments()
scrnaseq_transform = ln.Transform(
key="scrna-seq",
version="4.0.0",
type="pipeline",
reference="https://github.com/nf-core/scrnaseq",
).save()
run = ln.Run(transform=scrnaseq_transform).save()
register_pipeline_io(args.input, args.output, run)
register_pipeline_metadata(args.output, run)
!python register_scrnaseq_run.py --input scrnaseq_input --output scrnaseq_output
Show code cell output
→ connected lamindb: testuser1/test-nextflow
/home/runner/work/nf-lamin/nf-lamin/docs/register_scrnaseq_run.py:77: DeprecationWarning: `type` argument of transform was renamed to `kind` and will be removed in a future release.
scrnaseq_transform = ln.Transform(
! folder is outside existing storage location, will copy files from scrnaseq_input to /home/runner/work/nf-lamin/nf-lamin/docs/test-nextflow/scrnaseq_input
! data is a DataFrame, please use .from_dataframe()
Traceback (most recent call last):
File "/home/runner/work/nf-lamin/nf-lamin/docs/register_scrnaseq_run.py", line 85, in <module>
register_pipeline_metadata(args.output, run)
~~~~~~~~~~~~~~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^
File "/home/runner/work/nf-lamin/nf-lamin/docs/register_scrnaseq_run.py", line 59, in register_pipeline_metadata
artifact = ln.Artifact(
~~~~~~~~~~~^
matching_files[0],
^^^^^^^^^^^^^^^^^^
...<2 lines>...
run=False,
^^^^^^^^^^
).save()
^
File "/opt/hostedtoolcache/Python/3.14.3/x64/lib/python3.14/site-packages/lamindb/models/artifact.py", line 1639, in __init__
raise FieldValidationError(
f"Only {valid_keywords} can be passed, you passed: {kwargs}"
)
lamindb.errors.FieldValidationError: Only path, key, description, kind, features, schema, revises, overwrite_versions, run, storage, branch, space, skip_hash_lookup can be passed, you passed: {'visibility': 0}
Data lineage¶
The output data could now be accessed (in a different notebook/script) for analysis with full lineage.
matrix_af = ln.Artifact.get(key__icontains="filtered_count_matrix.h5ad")
matrix_af.view_lineage()
Show code cell output
View transforms & runs on the hub¶
# clean up the test instance:
!rm -rf test-nextflow
!lamin delete --force test-nextflow
