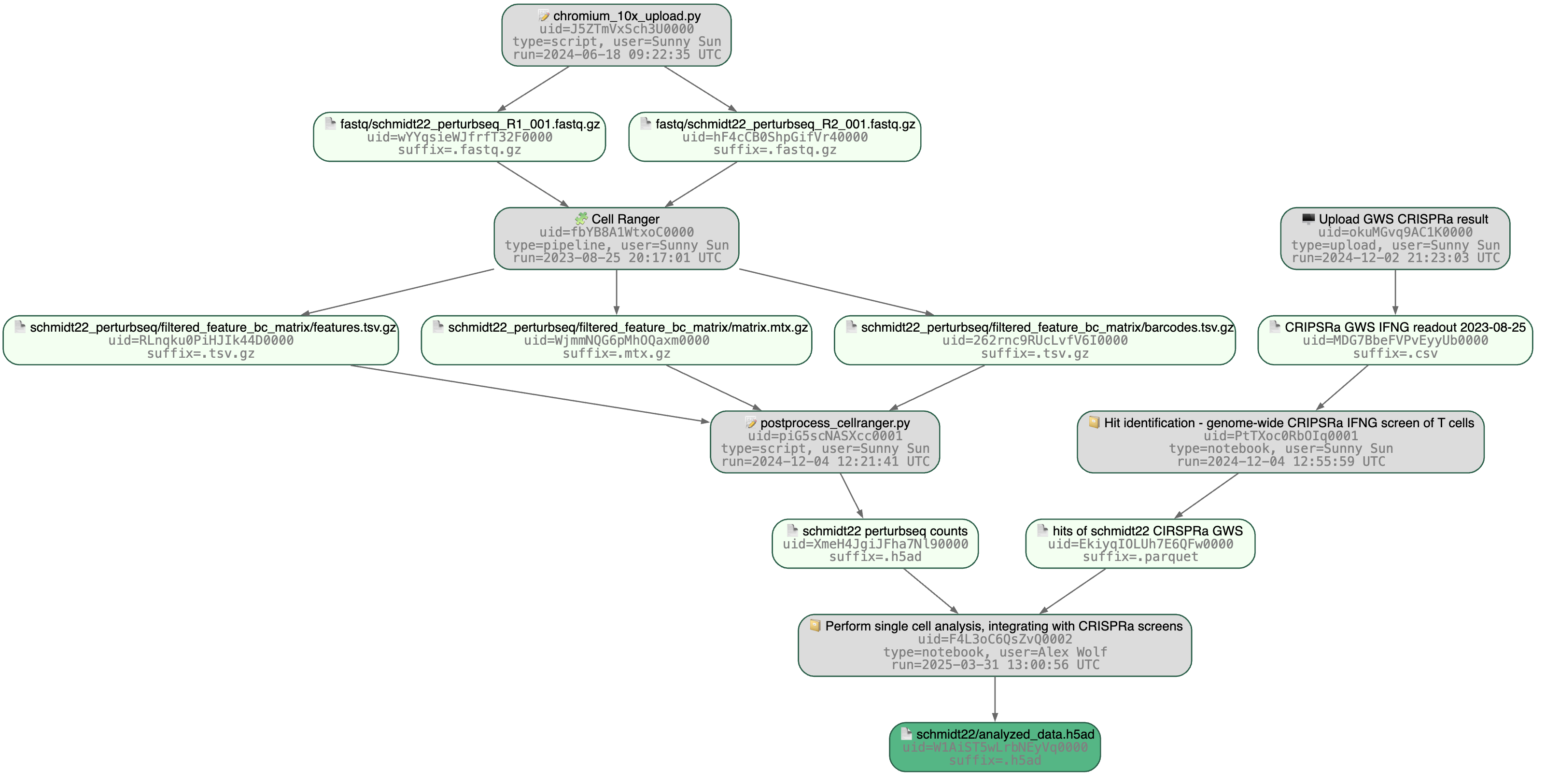
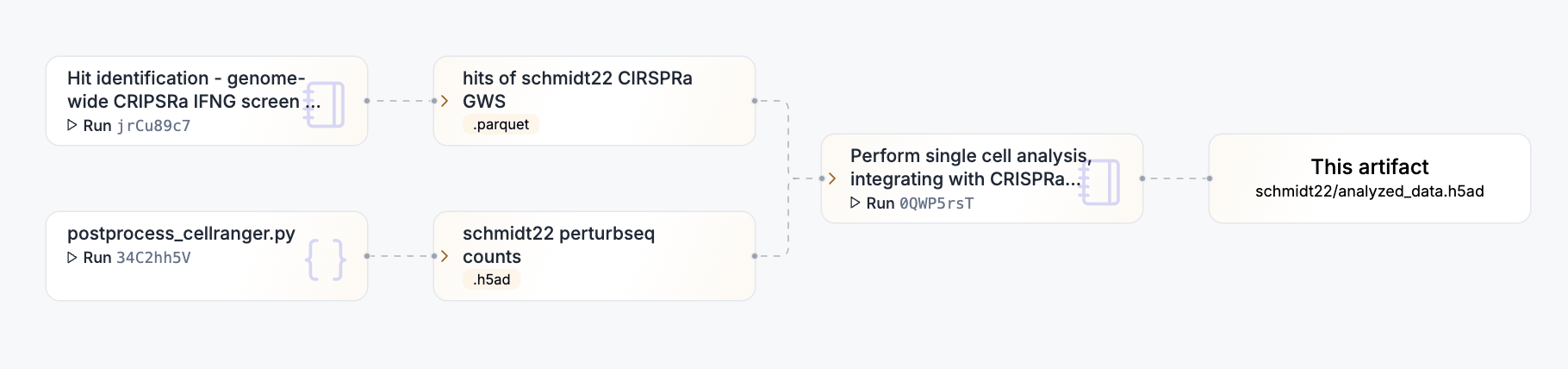
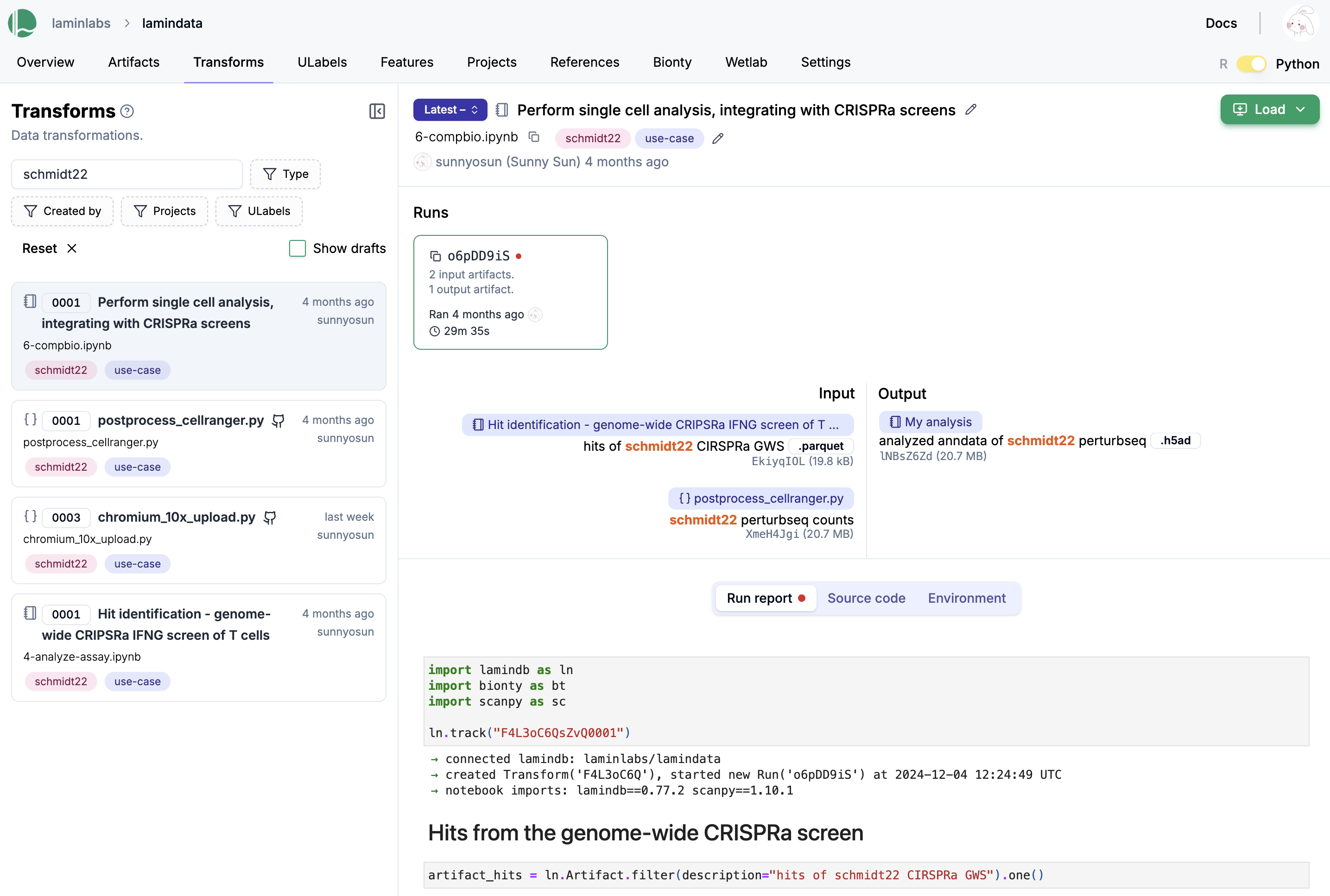
Tutorial 
 ¶
¶
LaminDB provides a framework to transform datasets into more useful representations: validated & queryable datasets, machine learning models, and analytical insights. The transformations can be notebooks, scripts, pipelines, or functions.
The metadata involved in this process are stored in a LaminDB instance, a database that manages datasets in storage. For the following walk through LaminDB’s core features, we’ll be working with a local instance.
Track a notebook or script¶
If you don’t have a LaminDB instance, create one using the shell:
!lamin init --storage ./lamindb-tutorial --modules bionty
Show code cell output
→ initialized lamindb: anonymous/lamindb-tutorial
Via the R shell
library(laminr)
lc <- import_module("lamin_cli")
lc$init(storage = "./lamin-tutorial", modules = "bionty")
What else can I configure during setup?
You can pass a cloud storage location to
--storage(S3, GCP, R2, HF, etc.)--storage s3://my-bucket
Instead of the default SQLite database pass a Postgres connection string to
--db:--db postgresql://<user>:<pwd>@<hostname>:<port>/<dbname>
Instead of a default instance name derived from the storage location, provide a custom name:
--name my-name
Mount additional schema modules:
--modules bionty,pertdb,custom1
For more info, see Install & setup .
Let’s now track the notebook that’s being run.
import lamindb as ln
ln.track() # track the current notebook or script
library(laminr)
ln <- import_module("lamindb")
ln$track() # track the current notebook or script
Show code cell outputs
→ connected lamindb: anonymous/lamindb-tutorial
→ created Transform('pkNMWqoQglTu0000', key='tutorial.ipynb'), started new Run('dHStEMKvYsWUiV3O') at 2026-02-24 15:35:24 UTC
→ notebook imports: anndata==0.12.10 bionty==2.2.1 lamindb==2.2.1
• recommendation: to identify the notebook across renames, pass the uid: ln.track("pkNMWqoQglTu")
By calling track(), the notebook gets automatically linked as the source of all data that’s about to be saved! You can see all your transforms and their runs in the Transform and Run registries.
ln.Transform.to_dataframe()
ln$Transform$to_dataframe()
Show code cell outputs
| uid | key | description | kind | source_code | hash | reference | reference_type | version_tag | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | environment_id | plan_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||||
| 1 | pkNMWqoQglTu0000 | tutorial.ipynb | Tutorial [
ln$Run$to_dataframe()
Show code cell outputs
| uid | name | description | entrypoint | started_at | finished_at | params | reference | reference_type | cli_args | ... | created_at | branch_id | created_on_id | space_id | transform_id | report_id | environment_id | plan_id | created_by_id | initiated_by_run_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 1 | dHStEMKvYsWUiV3O | None | None | None | 2026-02-24 15:35:24.182049+00:00 | None | None | None | None | None | ... | 2026-02-24 15:35:24.182000+00:00 | 1 | 1 | 1 | 1 | None | None | None | 1 | None |
1 rows × 21 columns
What happened under the hood?
The full run environment and imported package versions of current notebook were detected
Notebook metadata was detected and stored in a
Transformrecord with a unique idRun metadata was detected and stored in a
Runrecord with a unique id
The Transform registry stores data transformations: scripts, notebooks, pipelines, functions.
The Run registry stores executions of transforms. Many runs can be linked to the same transform if executed with different context (time, user, input data, etc.).
How do I track a pipeline instead of a notebook?
Leverage a pipeline integration, see: Manage computational pipelines. Or manually add code as seen below.
transform = ln.Transform(key="my_pipeline", version="1.2.0")
ln.track(transform)
Why should I care about tracking notebooks?
Because of interactivity & humans are in the loop, most mistakes happen when using notebooks.
track() makes notebooks & derived results reproducible & auditable, enabling to learn from mistakes.
This is important as much insight generated from biological data is driven by computational biologists interacting with it. An early blog post on this is here.
Is this compliant with OpenLineage?
Yes. What OpenLineage calls a “job”, LaminDB calls a “transform”. What OpenLineage calls a “run”, LaminDB calls a “run”.
Manage artifacts¶
The Artifact class manages datasets & models that are stored as files, folders, or arrays. Artifact is a registry to manage search, queries, validation & storage access.
You can register data objects (DataFrame, AnnData, …) and files or folders in local storage, AWS S3 (s3://), Google Cloud (gs://), Hugging Face (hf://), or any other file system supported by fsspec.
Create an artifact¶
Let’s first look at an exemplary dataframe.
df = ln.examples.datasets.mini_immuno.get_dataset1(with_typo=True)
df
df <- ln$examples$datasets$mini_immuno$get_dataset1(with_typo = TRUE)
df
Show code cell outputs
| ENSG00000153563 | ENSG00000010610 | ENSG00000170458 | perturbation | sample_note | cell_type_by_expert | cell_type_by_model | assay_oid | concentration | treatment_time_h | donor | donor_ethnicity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sample1 | 1 | 3 | 5 | DMSO | was ok | B cell | B cell | EFO:0008913 | 0.1% | 24 | D0001 | [Chinese, Singaporean Chinese] |
| sample2 | 2 | 4 | 6 | IFNJ | looks naah | CD8-positive, alpha-beta T cell | T cell | EFO:0008913 | 200 nM | 24 | D0002 | [Chinese, Han Chinese] |
| sample3 | 3 | 5 | 7 | DMSO | pretty! 🤩 | CD8-positive, alpha-beta T cell | T cell | EFO:0008913 | 0.1% | 6 | None | [Chinese] |
This is how you create an artifact from a dataframe.
artifact = ln.Artifact.from_dataframe(df, key="my_datasets/rnaseq1.parquet").save()
artifact.describe()
artifact <- ln$Artifact$from_dataframe(df, key = "my_datasets/rnaseq1.parquet")$save()
artifact$describe()
Show code cell outputs
Artifact: my_datasets/rnaseq1.parquet (0000) ├── uid: 2COV0ZySwwZ6n3U00000 run: dHStEMK (tutorial.ipynb) │ kind: dataset otype: DataFrame │ hash: 0E5-eyY-mQBw83oqN20Wjw size: 10.1 KB │ branch: main space: all │ created_at: 2026-02-24 15:35:25 UTC created_by: anonymous │ n_observations: 3 └── storage/path: /home/runner/work/lamin-docs/lamin-docs/docs/lamindb-tutorial/.lamindb/2COV0ZySwwZ6n3U00000.parquet
Access artifacts¶
Get the artifact by key.
artifact = ln.Artifact.get(key="my_datasets/rnaseq1.parquet")
artifact <- ln$Artifact$get(key = "my_datasets/rnaseq1.parquet")
And this is how you load it back into memory.
artifact.load()
artifact$load()
Show code cell outputs
| ENSG00000153563 | ENSG00000010610 | ENSG00000170458 | perturbation | sample_note | cell_type_by_expert | cell_type_by_model | assay_oid | concentration | treatment_time_h | donor | donor_ethnicity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sample1 | 1 | 3 | 5 | DMSO | was ok | B cell | B cell | EFO:0008913 | 0.1% | 24 | D0001 | [Chinese, Singaporean Chinese] |
| sample2 | 2 | 4 | 6 | IFNJ | looks naah | CD8-positive, alpha-beta T cell | T cell | EFO:0008913 | 200 nM | 24 | D0002 | [Chinese, Han Chinese] |
| sample3 | 3 | 5 | 7 | DMSO | pretty! 🤩 | CD8-positive, alpha-beta T cell | T cell | EFO:0008913 | 0.1% | 6 | None | [Chinese] |
Typically your artifact is in a cloud storage location. To get a local file path to it, call cache().
artifact.cache()
artifact$cache()
Show code cell outputs
PosixUPath('/home/runner/work/lamin-docs/lamin-docs/docs/lamindb-tutorial/.lamindb/2COV0ZySwwZ6n3U00000.parquet')
If the data is large, you might not want to cache but stream it via open(). For more on this, see: Stream datasets from storage .
Trace data lineage¶
You can understand where an artifact comes from by looking at its Transform & Run records.
artifact.transform
artifact$transform
Show code cell outputs
Transform(uid='pkNMWqoQglTu0000', version_tag=None, is_latest=True, key='tutorial.ipynb', description='Tutorial [](https://github.com/laminlabs/lamin-docs/blob/main/docs/tutorial.md)', kind='notebook', hash=None, reference=None, reference_type=None, environment=None, plan=None, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, created_at=2026-02-24 15:35:24 UTC, is_locked=False)
artifact.run
artifact$run
Show code cell outputs
Run(uid='dHStEMKvYsWUiV3O', name=None, description=None, entrypoint=None, started_at=2026-02-24 15:35:24 UTC, finished_at=None, params=None, reference=None, reference_type=None, cli_args=None, branch_id=1, created_on_id=1, space_id=1, transform_id=1, report_id=None, environment_id=None, plan_id=None, created_by_id=1, initiated_by_run_id=None, created_at=2026-02-24 15:35:24 UTC, is_locked=False)
Or visualize deeper data lineage with the view_lineage() method. Here we’re only one step deep.
artifact.view_lineage()
artifact$view_lineage()
Show code cell outputs
! calling anonymously, will miss private instances
Show me a more interesting example, please!
Once you’re done, at the end of your notebook or script, call finish(). Here, we’re not yet done so we’re commenting it out.
# ln.finish() # mark run as finished, save execution report, source code & environment
# ln$finish() # mark run as finished, save execution report, source code & environment
Saving reports when using R
If you did not use RStudio’s notebook mode, you have to render an HTML externally.
Render the notebook to HTML via one of:
In RStudio, click the “Knit” button
From the command line, run
Rscript -e 'rmarkdown::render("tutorial.Rmd")'
Use the
rmarkdownpackage in Rrmarkdown::render("tutorial.Rmd")
Save it via one of:
the
save()command in thelamin_climodule from Rlc <- import_module("lamin_cli") lc$save("tutorial.Rmd")
the
laminCLIlamin save tutorial.Rmd
Here is how a notebook looks on the hub.
To create a new version of a notebook or script, run lamin load on the terminal, e.g.,
$ lamin load https://lamin.ai/laminlabs/lamindata/transform/13VINnFk89PE0004
→ notebook is here: mcfarland_2020_preparation.ipynb
Note that data lineage also helps to understand what a dataset is being used for, not only where it comes from. Many datasets are being used over and over for different purposes and it’s often useful to understand how.
Annotate an artifact¶
You can annotate artifacts with features and labels. Features are measurement dimensions (e.g. "organism", "temperature") and labels are measured categories (e.g. "human", "mouse").
Let’s annotate an artifact with a Record, a built-in universal label ontology.
# create & save a record
my_experiment = ln.Record(name="My experiment").save()
# annotate the artifact with a record
artifact.records.add(my_experiment)
# describe the artifact
artifact.describe()
# create & save a record
my_experiment <- ln$Record(name = "My experiment")$save()
# annotate the artifact with a record
artifact$records$add(my_experiment)
# describe the artifact
artifact$describe()
Show code cell outputs
Artifact: my_datasets/rnaseq1.parquet (0000) ├── uid: 2COV0ZySwwZ6n3U00000 run: dHStEMK (tutorial.ipynb) │ kind: dataset otype: DataFrame │ hash: 0E5-eyY-mQBw83oqN20Wjw size: 10.1 KB │ branch: main space: all │ created_at: 2026-02-24 15:35:25 UTC created_by: anonymous │ n_observations: 3 ├── storage/path: │ /home/runner/work/lamin-docs/lamin-docs/docs/lamindb-tutorial/.lamindb/2COV0ZySwwZ6n3U00000.parquet └── Labels └── .records Record My experiment
This is how you query artifacts based on the annotation.
ln.Artifact.filter(records=my_experiment).to_dataframe()
ln$Artifact$filter(records = my_experiment)$to_dataframe()
Show code cell outputs
| uid | key | description | suffix | kind | otype | size | hash | n_files | n_observations | ... | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | storage_id | run_id | schema_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 1 | 2COV0ZySwwZ6n3U00000 | my_datasets/rnaseq1.parquet | None | .parquet | dataset | DataFrame | 10354 | 0E5-eyY-mQBw83oqN20Wjw | None | 3 | ... | True | False | 2026-02-24 15:35:25.740000+00:00 | 1 | 1 | 1 | 1 | 1 | None | 1 |
1 rows × 21 columns
You can also annotate with labels from other registries, e.g., the biological ontologies in bionty.
import bionty as bt
# create a cell type label from the source ontology
cell_type = bt.CellType.from_source(name="effector T cell").save()
# annotate the artifact with a cell type
artifact.cell_types.add(cell_type)
# describe the artifact
artifact.describe()
bt <- import_module("bionty")
# create a cell type label from the source ontology
cell_type <- bt$CellType$from_source(name = "effector T cell")$save()
# annotate the artifact with a cell type
artifact$cell_types$add(cell_type)
# describe the artifact
artifact$describe()
Show code cell outputs
Artifact: my_datasets/rnaseq1.parquet (0000) ├── uid: 2COV0ZySwwZ6n3U00000 run: dHStEMK (tutorial.ipynb) │ kind: dataset otype: DataFrame │ hash: 0E5-eyY-mQBw83oqN20Wjw size: 10.1 KB │ branch: main space: all │ created_at: 2026-02-24 15:35:25 UTC created_by: anonymous │ n_observations: 3 ├── storage/path: │ /home/runner/work/lamin-docs/lamin-docs/docs/lamindb-tutorial/.lamindb/2COV0ZySwwZ6n3U00000.parquet └── Labels └── .records Record My experiment .cell_types bionty.CellType effector T cell
This is how you query artifacts by cell type annotations.
ln.Artifact.filter(cell_types=cell_type).to_dataframe()
ln$Artifact$filter(cell_types = cell_type)$to_dataframe()
Show code cell outputs
| uid | key | description | suffix | kind | otype | size | hash | n_files | n_observations | ... | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | storage_id | run_id | schema_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 1 | 2COV0ZySwwZ6n3U00000 | my_datasets/rnaseq1.parquet | None | .parquet | dataset | DataFrame | 10354 | 0E5-eyY-mQBw83oqN20Wjw | None | 3 | ... | True | False | 2026-02-24 15:35:25.740000+00:00 | 1 | 1 | 1 | 1 | 1 | None | 1 |
1 rows × 21 columns
If you want to annotate by non-categorical metadata or indicate the feature for a label, annotate via features.
# define the "temperature" & "experiment" features
ln.Feature(name="temperature", dtype=float).save()
ln.Feature(name="experiment", dtype=ln.Record).save()
# annotate the artifact
artifact.features.add_values({"temperature": 21.6, "experiment": "My experiment"})
# describe the artifact
artifact.describe()
# define the "temperature" & "experiment" features
ln$Feature(name = "temperature", dtype = "float")$save()
ln$Feature(name = "experiment", dtype = "ln"$Record)$save()
# annotate the artifact
artifact$features$add_values(list(temperature = 21.6, experiment = "My experiment"))
# describe the artifact
artifact$describe()
Show code cell outputs
Artifact: my_datasets/rnaseq1.parquet (0000) ├── uid: 2COV0ZySwwZ6n3U00000 run: dHStEMK (tutorial.ipynb) │ kind: dataset otype: DataFrame │ hash: 0E5-eyY-mQBw83oqN20Wjw size: 10.1 KB │ branch: main space: all │ created_at: 2026-02-24 15:35:25 UTC created_by: anonymous │ n_observations: 3 ├── storage/path: │ /home/runner/work/lamin-docs/lamin-docs/docs/lamindb-tutorial/.lamindb/2COV0ZySwwZ6n3U00000.parquet ├── Features │ └── experiment Record My experiment │ temperature float 21.6 └── Labels └── .records Record My experiment .cell_types bionty.CellType effector T cell
This is how you query artifacts by features.
ln.Artifact.filter(temperature=21.6).to_dataframe()
ln$Artifact$filter(temperature = 21.6)$to_dataframe()
Show code cell outputs
| uid | key | description | suffix | kind | otype | size | hash | n_files | n_observations | ... | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | storage_id | run_id | schema_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 1 | 2COV0ZySwwZ6n3U00000 | my_datasets/rnaseq1.parquet | None | .parquet | dataset | DataFrame | 10354 | 0E5-eyY-mQBw83oqN20Wjw | None | 3 | ... | True | False | 2026-02-24 15:35:25.740000+00:00 | 1 | 1 | 1 | 1 | 1 | None | 1 |
1 rows × 21 columns
Validate an artifact¶
Validated datasets are more re-usable by analysts and machine learning models. You can define what a valid artifact should look like by defining a schema.
In lamindb, validation also means annotation with the validated metadata which increases the findability of a dataset.
Can you give me examples for what findability and usability means?
Findability: Which datasets measured expression of cell marker
CD14? Which characterized cell lineK562? Which have a test & train split? Etc.Usability: Are there typos in feature names? Are there typos in labels? Are types and units of features consistent? Etc.
import bionty as bt # <-- use bionty to access registries with imported public ontologies
# define a few more valid labels
ln.Record(name="DMSO").save()
ln.Record(name="IFNG").save()
# define a few more valid features
ln.Feature(name="perturbation", dtype=ln.Record).save()
ln.Feature(name="cell_type_by_model", dtype=bt.CellType).save()
ln.Feature(name="cell_type_by_expert", dtype=bt.CellType).save()
ln.Feature(name="assay_oid", dtype=bt.ExperimentalFactor.ontology_id).save()
ln.Feature(name="donor", dtype=str, nullable=True).save()
ln.Feature(name="sample_note", dtype=str, nullable=True).save()
ln.Feature(name="concentration", dtype=str).save()
ln.Feature(name="treatment_time_h", dtype="num", coerce=True).save()
# define a schema that merely enforces a feature identifier type
schema = ln.Schema(itype=ln.Feature).save()
bt <- import_module("bionty")
# define a few more valid labels
ln$Record(name = "DMSO")$save()
ln$Record(name = "IFNG")$save()
# define a few more valid features
ln$Feature(name = "perturbation", dtype = "ln"$Record)$save()
ln$Feature(name = "cell_type_by_model", dtype = "bt"$CellType)$save()
ln$Feature(name = "cell_type_by_expert", dtype = "bt"$CellType)$save()
ln$Feature(name = "assay_oid", dtype = "bt"$ExperimentalFactor$ontology_id)$save()
ln$Feature(name = "donor", dtype = "str", nullable = TRUE)$save()
ln$Feature(name = "sample_note", dtype = "str", nullable = TRUE)$save()
ln$Feature(name = "concentration", dtype = "str")$save()
ln$Feature(name = "treatment_time_h", dtype = "num", coerce = TRUE)$save()
# define a schema that merely enforces a feature identifier type
schema <- ln$Schema(itype = ln$Feature)$save()
If you pass a schema object to the Artifact constructor, the artifact will be validated & annotated. Let’s try this.
try:
artifact = ln.Artifact.from_dataframe(
df, key="my_datasets/rnaseq1.parquet", schema=schema
)
except ln.errors.ValidationError as error:
print(str(error))
try:
artifact = ln$Artifact$from_dataframe(
df, key = "my_datasets/rnaseq1.parquet", schema = schema
)
except ln$errors$ValidationError as error:
print(str(error))
Show code cell outputs
→ returning artifact with same hash: Artifact(uid='2COV0ZySwwZ6n3U00000', version_tag=None, is_latest=True, key='my_datasets/rnaseq1.parquet', description=None, suffix='.parquet', kind='dataset', otype='DataFrame', size=10354, hash='0E5-eyY-mQBw83oqN20Wjw', n_files=None, n_observations=3, branch_id=1, created_on_id=1, space_id=1, storage_id=1, run_id=1, schema_id=None, created_by_id=1, created_at=2026-02-24 15:35:25 UTC, is_locked=False); to track this artifact as an input, use: ln.Artifact.get()
→ loading artifact into memory for validation
! 4 terms not validated in feature 'columns': 'ENSG00000153563', 'ENSG00000010610', 'ENSG00000170458', 'donor_ethnicity'
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('columns')
! 1 term not validated in feature 'perturbation': 'IFNJ'
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('perturbation')
1 term not validated in feature 'perturbation': 'IFNJ'
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('perturbation')
Because there is a typo in the perturbation column, validation fails. Let’s fix it by making a new version.
Make a new version of an artifact¶
# fix the "IFNJ" typo
df["perturbation"] = df["perturbation"].cat.rename_categories({"IFNJ": "IFNG"})
# create a new version
artifact = ln.Artifact.from_dataframe(
df, key="my_datasets/rnaseq1.parquet", schema=schema
).save()
# see the annotations
artifact.describe()
# see all versions of the artifact
artifact.versions.to_dataframe()
# fix the "IFNJ" typo
dflist("perturbation") = dflist("perturbation")$cat$rename_categories(list(IFNJ = "IFNG"))
# create a new version
artifact <- ln$Artifact$from_dataframe(
df, key = "my_datasets/rnaseq1.parquet", schema = schema
)$save()
# see the annotations
artifact$describe()
# see all versions of the artifact
artifact$versions$to_dataframe()
Show code cell outputs
→ creating new artifact version for key 'my_datasets/rnaseq1.parquet' in storage '/home/runner/work/lamin-docs/lamin-docs/docs/lamindb-tutorial'
→ loading artifact into memory for validation
! 4 terms not validated in feature 'columns': 'ENSG00000153563', 'ENSG00000010610', 'ENSG00000170458', 'donor_ethnicity'
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('columns')
Artifact: my_datasets/rnaseq1.parquet (0001) ├── uid: 2COV0ZySwwZ6n3U00001 run: dHStEMK (tutorial.ipynb) │ kind: dataset otype: DataFrame │ hash: o2OEY7jSQiUOx4hxqrxaMw size: 10.1 KB │ branch: main space: all │ created_at: 2026-02-24 15:35:32 UTC created_by: anonymous │ n_observations: 3 ├── storage/path: │ /home/runner/work/lamin-docs/lamin-docs/docs/lamindb-tutorial/.lamindb/2COV0ZySwwZ6n3U00001.parquet ├── Dataset features │ └── columns (8) │ assay_oid bionty.ExperimentalFactor.ontology… EFO:0008913 │ cell_type_by_expert bionty.CellType B cell, CD8-positive, alpha-beta T cell │ cell_type_by_model bionty.CellType B cell, T cell │ concentration str │ donor str │ perturbation Record DMSO, IFNG │ sample_note str │ treatment_time_h num └── Labels └── .records Record DMSO, IFNG .cell_types bionty.CellType T cell, B cell, CD8-positive, alpha-be… .experimental_factors bionty.ExperimentalFactor single-cell RNA sequencing
| uid | key | description | suffix | kind | otype | size | hash | n_files | n_observations | ... | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | storage_id | run_id | schema_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 2 | 2COV0ZySwwZ6n3U00001 | my_datasets/rnaseq1.parquet | None | .parquet | dataset | DataFrame | 10354 | o2OEY7jSQiUOx4hxqrxaMw | None | 3 | ... | True | False | 2026-02-24 15:35:32.087000+00:00 | 1 | 1 | 1 | 1 | 1 | 1.0 | 1 |
| 1 | 2COV0ZySwwZ6n3U00000 | my_datasets/rnaseq1.parquet | None | .parquet | dataset | DataFrame | 10354 | 0E5-eyY-mQBw83oqN20Wjw | None | 3 | ... | False | False | 2026-02-24 15:35:25.740000+00:00 | 1 | 1 | 1 | 1 | 1 | NaN | 1 |
2 rows × 21 columns
The content of the dataset is now validated and the dataset is richly annotated and queryable by all entities that you defined.
Can I also create new versions without passing key?
That works, too, you can use revises:
artifact_v1 = ln.Artifact.from_dataframe(df, description="Just a description").save()
# below revises artifact_v1
artifact_v2 = ln.Artifact.from_dataframe(df_updated, revises=artifact_v1).save()
The good thing about passing revises: Artifact is that you don’t need to worry about coming up with naming conventions for paths.
The good thing about versioning based on key is that it’s how all data versioning tools are doing it.
Query & search registries¶
We’ve already seen a few queries. Let’s now walk through the topic systematically.
To get an overview over all artifacts in your instance, call df.
ln.Artifact.to_dataframe()
ln$Artifact$to_dataframe()
Show code cell outputs
| uid | key | description | suffix | kind | otype | size | hash | n_files | n_observations | ... | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | storage_id | run_id | schema_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 2 | 2COV0ZySwwZ6n3U00001 | my_datasets/rnaseq1.parquet | None | .parquet | dataset | DataFrame | 10354 | o2OEY7jSQiUOx4hxqrxaMw | None | 3 | ... | True | False | 2026-02-24 15:35:32.087000+00:00 | 1 | 1 | 1 | 1 | 1 | 1.0 | 1 |
| 1 | 2COV0ZySwwZ6n3U00000 | my_datasets/rnaseq1.parquet | None | .parquet | dataset | DataFrame | 10354 | 0E5-eyY-mQBw83oqN20Wjw | None | 3 | ... | False | False | 2026-02-24 15:35:25.740000+00:00 | 1 | 1 | 1 | 1 | 1 | NaN | 1 |
2 rows × 21 columns
The Artifact registry additionally supports seeing all feature annotations of an artifact.
ln.Artifact.to_dataframe(features=True)
ln$Artifact$to_dataframe(features = TRUE)
Show code cell outputs
→ queried for all categorical features of dtypes Record or ULabel and non-categorical features: (7) ['temperature', 'experiment', 'perturbation', 'donor', 'sample_note', 'concentration', 'treatment_time_h']
| uid | key | temperature | experiment | perturbation | |
|---|---|---|---|---|---|
| id | |||||
| 2 | 2COV0ZySwwZ6n3U00001 | my_datasets/rnaseq1.parquet | NaN | NaN | {IFNG, DMSO} |
| 1 | 2COV0ZySwwZ6n3U00000 | my_datasets/rnaseq1.parquet | 21.6 | My experiment | NaN |
LaminDB’s central classes are registries that store records (SQLRecord objects). If you want to see the fields of a registry, look at the class or auto-complete.
ln.Artifact
ln$Artifact
Show code cell outputs
lamindb.models.artifact.Artifact
Each registry is a table in the relational schema of the underlying database. With view(), you can see the latest records in the database.
ln.view()
ln$view()
Show code cell outputs
****************
* module: core *
****************
Artifact
| uid | key | description | suffix | kind | otype | size | hash | n_files | n_observations | ... | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | storage_id | run_id | schema_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 2 | 2COV0ZySwwZ6n3U00001 | my_datasets/rnaseq1.parquet | None | .parquet | dataset | DataFrame | 10354 | o2OEY7jSQiUOx4hxqrxaMw | None | 3 | ... | True | False | 2026-02-24 15:35:32.087000+00:00 | 1 | 1 | 1 | 1 | 1 | 1.0 | 1 |
| 1 | 2COV0ZySwwZ6n3U00000 | my_datasets/rnaseq1.parquet | None | .parquet | dataset | DataFrame | 10354 | 0E5-eyY-mQBw83oqN20Wjw | None | 3 | ... | False | False | 2026-02-24 15:35:25.740000+00:00 | 1 | 1 | 1 | 1 | 1 | NaN | 1 |
2 rows × 21 columns
Feature
| uid | name | _dtype_str | unit | description | array_rank | array_size | array_shape | synonyms | default_value | ... | coerce | is_locked | is_type | created_at | branch_id | created_on_id | space_id | created_by_id | run_id | type_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 10 | lLTumMunMic7 | treatment_time_h | num | None | None | 0 | 0 | None | None | None | ... | True | False | False | 2026-02-24 15:35:29.223000+00:00 | 1 | 1 | 1 | 1 | 1 | None |
| 9 | AFE4PxxxbMBh | concentration | str | None | None | 0 | 0 | None | None | None | ... | None | False | False | 2026-02-24 15:35:29.219000+00:00 | 1 | 1 | 1 | 1 | 1 | None |
| 8 | 2xHuAdbMbCag | sample_note | str | None | None | 0 | 0 | None | None | None | ... | None | False | False | 2026-02-24 15:35:29.215000+00:00 | 1 | 1 | 1 | 1 | 1 | None |
| 7 | v03Zr0xzqxqC | donor | str | None | None | 0 | 0 | None | None | None | ... | None | False | False | 2026-02-24 15:35:29.210000+00:00 | 1 | 1 | 1 | 1 | 1 | None |
| 6 | JgGBID4mHHix | assay_oid | cat[bionty.ExperimentalFactor.ontology_id] | None | None | 0 | 0 | None | None | None | ... | None | False | False | 2026-02-24 15:35:29.206000+00:00 | 1 | 1 | 1 | 1 | 1 | None |
| 5 | 5W3emlB4aLIL | cell_type_by_expert | cat[bionty.CellType] | None | None | 0 | 0 | None | None | None | ... | None | False | False | 2026-02-24 15:35:29.202000+00:00 | 1 | 1 | 1 | 1 | 1 | None |
| 4 | SmThoClxgyfu | cell_type_by_model | cat[bionty.CellType] | None | None | 0 | 0 | None | None | None | ... | None | False | False | 2026-02-24 15:35:29.198000+00:00 | 1 | 1 | 1 | 1 | 1 | None |
7 rows × 21 columns
JsonValue
| value | hash | is_locked | created_at | branch_id | created_on_id | space_id | created_by_id | run_id | feature_id | |
|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||
| 1 | 21.6 | XftFE5byhwPHY-11WjfNAw | False | 2026-02-24 15:35:29.104000+00:00 | 1 | 1 | 1 | 1 | 1 | 1 |
Record
| uid | name | description | reference | reference_type | extra_data | is_locked | is_type | created_at | branch_id | created_on_id | space_id | created_by_id | type_id | schema_id | run_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||
| 3 | 7W7d8xSnsjOIFNAY | IFNG | None | None | None | None | False | False | 2026-02-24 15:35:29.189000+00:00 | 1 | 1 | 1 | 1 | None | None | 1 |
| 2 | a5DCaGhZPWYyF5Ji | DMSO | None | None | None | None | False | False | 2026-02-24 15:35:29.184000+00:00 | 1 | 1 | 1 | 1 | None | None | 1 |
| 1 | YdCBZzIDFyhMGvdX | My experiment | None | None | None | None | False | False | 2026-02-24 15:35:26.604000+00:00 | 1 | 1 | 1 | 1 | None | None | 1 |
Run
| uid | name | description | entrypoint | started_at | finished_at | params | reference | reference_type | cli_args | ... | created_at | branch_id | created_on_id | space_id | transform_id | report_id | environment_id | plan_id | created_by_id | initiated_by_run_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 1 | dHStEMKvYsWUiV3O | None | None | None | 2026-02-24 15:35:24.182049+00:00 | None | None | None | None | None | ... | 2026-02-24 15:35:24.182000+00:00 | 1 | 1 | 1 | 1 | None | None | None | 1 | None |
1 rows × 21 columns
Schema
| uid | name | description | n_members | coerce | flexible | itype | otype | hash | minimal_set | ... | maximal_set | is_locked | is_type | created_at | branch_id | created_on_id | space_id | created_by_id | run_id | type_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 2 | 5we5Kvp3E9AayEr7 | None | None | 8.0 | None | False | Feature | None | V3yLoat04fZMXmIp8VmHxw | True | ... | False | False | False | 2026-02-24 15:35:32.105000+00:00 | 1 | 1 | 1 | 1 | 1 | None |
| 1 | 0000000000000000 | None | None | NaN | None | True | Feature | None | kMi7B_N88uu-YnbTLDU-DA | True | ... | False | False | False | 2026-02-24 15:35:29.226000+00:00 | 1 | 1 | 1 | 1 | 1 | None |
2 rows × 21 columns
Storage
| uid | root | description | type | region | instance_uid | is_locked | created_at | branch_id | created_on_id | space_id | created_by_id | run_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||
| 1 | VLuWYwO8Gpy1 | /home/runner/work/lamin-docs/lamin-docs/docs/l... | None | local | None | 5WCoFhciyw3n | False | 2026-02-24 15:35:21.870000+00:00 | 1 | 1 | 1 | 1 | None |
Transform
| uid | key | description | kind | source_code | hash | reference | reference_type | version_tag | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | environment_id | plan_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||||
| 1 | pkNMWqoQglTu0000 | tutorial.ipynb | Tutorial [
transform = ln.Transform.get(key="tutorial.ipynb")
# get a set of records by filtering for a directory (LaminDB treats directories like AWS S3, as the prefix of the storage key)
ln.Artifact.filter(key__startswith="my_datasets/").to_dataframe()
# query all artifacts ingested from a transform
artifacts = ln.Artifact.filter(transform=transform).all()
# query all artifacts ingested from a notebook with "tutor" in the description
artifacts = ln.Artifact.filter(
transform__description__icontains="tutor",
).all()
# get a single record (here the current notebook)
transform <- ln$Transform$get(key = "tutorial.ipynb")
# get a set of records by filtering for a directory (LaminDB treats directories like AWS S3, as the prefix of the storage key)
ln$Artifact$filter(key__startswith = "my_datasets/")$to_dataframe()
# query all artifacts ingested from a transform
artifacts <- ln$Artifact$filter(transform = transform)$all()
# query all artifacts ingested from a notebook with "tutor" in the description
artifacts <- ln$Artifact$filter(
transform__description__icontains = "tutor",
)$all()
What does a double underscore mean?
For any field, the double underscore defines a comparator, e.g.,
name__icontains="Martha":namecontains"Martha"when ignoring casename__startswith="Martha":namestarts with"Marthaname__in=["Martha", "John"]:nameis"John"or"Martha"
For more info, see: Query & search registries .
Can I chain filters and searches?
Yes: ln.Artifact.filter(suffix=".jpg").search("my image")
The class methods search and lookup help with approximate matches.
# search artifacts
ln.Artifact.search("iris").to_dataframe().head()
# search transforms
ln.Transform.search("tutor").to_dataframe()
# look up records with auto-complete
records = ln.Record.lookup()
# search artifacts
ln$Artifact$search("iris")$to_dataframe()$head()
# search transforms
ln$Transform$search("tutor")$to_dataframe()
# look up records with auto-complete
records <- ln$Record$lookup()
Show me a screenshot
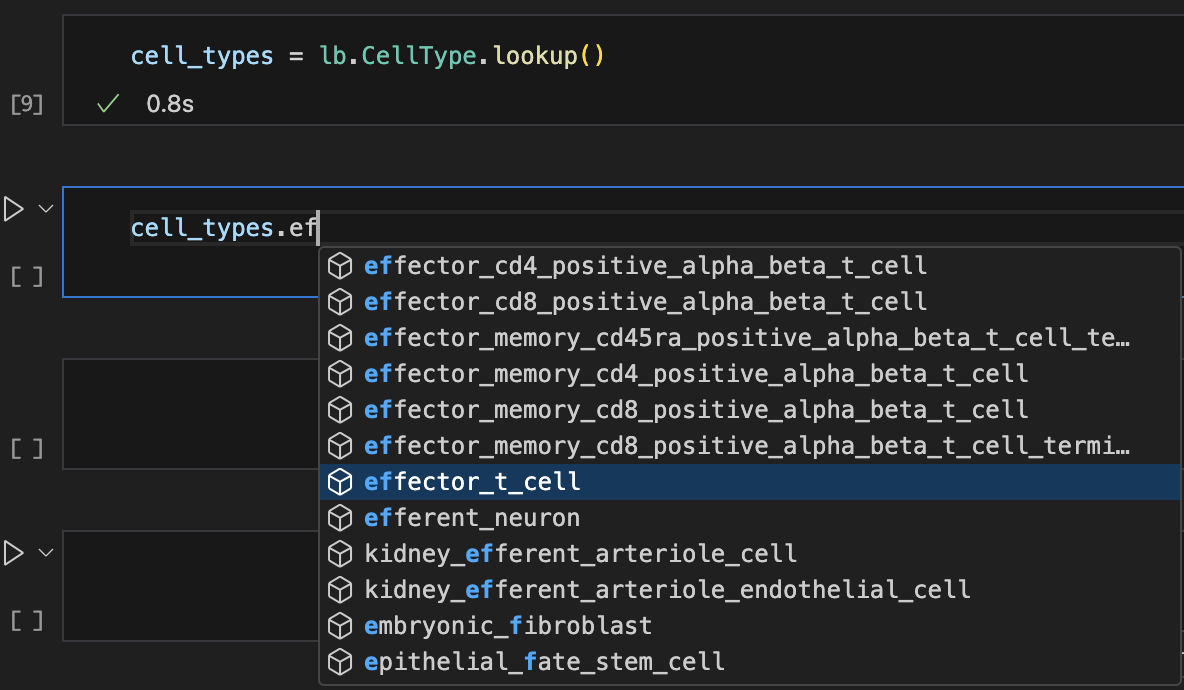
For more info, see: Query & search registries .
Manage files & folders¶
Let’s look at a folder in the cloud that contains 3 sub-folders storing images & metadata of Iris flowers, generated in 3 subsequent studies.
# we use anon=True here in case no aws credentials are configured
ln.UPath("s3://lamindata/iris_studies", anon=True).view_tree()
# we use anon = TRUE here in case no aws credentials are configured
ln$UPath("s3://lamindata/iris_studies", anon = TRUE)$view_tree()
Show code cell outputs
3 sub-directories & 151 files with suffixes '.csv', '.jpg'
s3://lamindata/iris_studies
├── study0_raw_images/
│ ├── iris-0337d20a3b7273aa0ddaa7d6afb57a37a759b060e4401871db3cefaa6adc068d.jpg
│ ├── iris-0797945218a97d6e5251b4758a2ba1b418cbd52ce4ef46a3239e4b939bd9807b.jpg
│ ├── iris-0f133861ea3fe1b68f9f1b59ebd9116ff963ee7104a0c4200218a33903f82444.jpg
│ ├── iris-0fec175448a23db03c1987527f7e9bb74c18cffa76ef003f962c62603b1cbb87.jpg
│ ├── iris-125b6645e086cd60131764a6bed12650e0f7f2091c8bbb72555c103196c01881.jpg
│ ├── iris-13dfaff08727abea3da8cfd8d097fe1404e76417fefe27ff71900a89954e145a.jpg
│ ├── iris-1566f7f5421eaf423a82b3c1cd1328f2a685c5ef87d8d8e710f098635d86d3d0.jpg
│ ├── iris-1804702f49c2c385f8b30913569aebc6dce3da52ec02c2c638a2b0806f16014e.jpg
│ ├── iris-318d451a8c95551aecfde6b55520f302966db0a26a84770427300780b35aa05a.jpg
│ ├── iris-3dec97fe46d33e194520ca70740e4c2e11b0ffbffbd0aec0d06afdc167ddf775.jpg
│ ├── iris-3eed72bc2511f619190ce79d24a0436fef7fcf424e25523cb849642d14ac7bcf.jpg
│ ├── iris-430fa45aad0edfeb5b7138ff208fdeaa801b9830a9eb68f378242465b727289a.jpg
│ ├── iris-4cc15cd54152928861ecbdc8df34895ed463403efb1571dac78e3223b70ef569.jpg
│ ├── iris-4febb88ef811b5ca6077d17ef8ae5dbc598d3f869c52af7c14891def774d73fa.jpg
│ ├── iris-590e7f5b8f4de94e4b82760919abd9684ec909d9f65691bed8e8f850010ac775.jpg
│ ├── iris-5a313749aa61e9927389affdf88dccdf21d97d8a5f6aa2bd246ca4bc926903ba.jpg
│ ├── iris-5b3106db389d61f4277f43de4953e660ff858d8ab58a048b3d8bf8d10f556389.jpg
│ ├── iris-5f4e8fffde2404cc30be275999fddeec64f8a711ab73f7fa4eb7667c8475c57b.jpg
│ ├── iris-68d83ad09262afb25337ccc1d0f3a6d36f118910f36451ce8a6600c77a8aa5bd.jpg
│ ├── iris-70069edd7ab0b829b84bb6d4465b2ca4038e129bb19d0d3f2ba671adc03398cc.jpg
│ ├── iris-7038aef1137814473a91f19a63ac7a55a709c6497e30efc79ca57cfaa688f705.jpg
│ ├── iris-74d1acf18cfacd0a728c180ec8e1c7b4f43aff72584b05ac6b7c59f5572bd4d4.jpg
│ ├── iris-7c3b5c5518313fc6ff2c27fcbc1527065cbb42004d75d656671601fa485e5838.jpg
│ ├── iris-7cf1ebf02b2cc31539ed09ab89530fec6f31144a0d5248a50e7c14f64d24fe6e.jpg
│ ├── iris-7dcc69fa294fe04767706c6f455ea6b31d33db647b08aab44b3cd9022e2f2249.jpg
│ ├── iris-801b7efb867255e85137bc1e1b06fd6cbab70d20cab5b5046733392ecb5b3150.jpg
│ ├── iris-8305dd2a080e7fe941ea36f3b3ec0aa1a195ad5d957831cf4088edccea9465e2.jpg
│ ├── iris-83f433381b755101b9fc9fbc9743e35fbb8a1a10911c48f53b11e965a1cbf101.jpg
│ ├── iris-874121a450fa8a420bdc79cc7808fd28c5ea98758a4b50337a12a009fa556139.jpg
│ ├── iris-8c216e1acff39be76d6133e1f549d138bf63359fa0da01417e681842210ea262.jpg
│ ├── iris-92c4268516ace906ad1ac44592016e36d47a8c72a51cacca8597ba9e18a8278b.jpg
│ ├── iris-95d7ec04b8158f0873fa4aab7b0a5ec616553f3f9ddd6623c110e3bc8298248f.jpg
│ ├── iris-9ce2d8c4f1eae5911fcbd2883137ba5542c87cc2fe85b0a3fbec2c45293c903e.jpg
│ ├── iris-9ee27633bb041ef1b677e03e7a86df708f63f0595512972403dcf5188a3f48f5.jpg
│ ├── iris-9fb8d691550315506ae08233406e8f1a4afed411ea0b0ac37e4b9cdb9c42e1ec.jpg
│ ├── iris-9ffe51c2abd973d25a299647fa9ccaf6aa9c8eecf37840d7486a061438cf5771.jpg
│ ├── iris-a2be5db78e5b603a5297d9a7eec4e7f14ef2cba0c9d072dc0a59a4db3ab5bb13.jpg
│ ├── iris-ad7da5f15e2848ca269f28cd1dc094f6f685de2275ceaebb8e79d2199b98f584.jpg
│ ├── iris-bc515e63b5a4af49db8c802c58c83db69075debf28c792990d55a10e881944d9.jpg
│ ├── iris-bd8d83096126eaa10c44d48dbad4b36aeb9f605f1a0f6ca929d3d0d492dafeb6.jpg
│ ├── iris-bdae8314e4385d8e2322abd8e63a82758a9063c77514f49fc252e651cbd79f82.jpg
│ ├── iris-c175cd02ac392ecead95d17049f5af1dcbe37851c3e42d73e6bb813d588ea70b.jpg
│ ├── iris-c31e6056c94b5cb618436fbaac9eaff73403fa1b87a72db2c363d172a4db1820.jpg
│ ├── iris-ca40bc5839ee2f9f5dcac621235a1db2f533f40f96a35e1282f907b40afa457d.jpg
│ ├── iris-ddb685c56cfb9c8496bcba0d57710e1526fff7d499536b3942d0ab375fa1c4a6.jpg
│ ├── iris-e437a7c7ad2bbac87fef3666b40c4de1251b9c5f595183eda90a8d9b1ef5b188.jpg
│ ├── iris-e7e0774289e2153cc733ff62768c40f34ac9b7b42e23c1abc2739f275e71a754.jpg
│ ├── iris-e9da6dd69b7b07f80f6a813e2222eae8c8f7c3aeaa6bcc02b25ea7d763bcf022.jpg
│ ├── iris-eb01666d4591b2e03abecef5a7ded79c6d4ecb6d1922382c990ad95210d55795.jpg
│ ├── iris-f6e4890dee087bd52e2c58ea4c6c2652da81809603ea3af561f11f8c2775c5f3.jpg
│ └── meta.csv
├── study1_raw_images/
│ ├── iris-0879d3f5b337fe512da1c7bf1d2bfd7616d744d3eef7fa532455a879d5cc4ba0.jpg
│ ├── iris-0b486eebacd93e114a6ec24264e035684cebe7d2074eb71eb1a71dd70bf61e8f.jpg
│ ├── iris-0ff5ba898a0ec179a25ca217af45374fdd06d606bb85fc29294291facad1776a.jpg
│ ├── iris-1175239c07a943d89a6335fb4b99a9fb5aabb2137c4d96102f10b25260ae523f.jpg
│ ├── iris-1289c57b571e8e98e4feb3e18a890130adc145b971b7e208a6ce5bad945b4a5a.jpg
│ ├── iris-12adb3a8516399e27ff1a9d20d28dca4674836ed00c7c0ae268afce2c30c4451.jpg
│ ├── iris-17ac8f7b5734443090f35bdc531bfe05b0235b5d164afb5c95f9d35f13655cf3.jpg
│ ├── iris-2118d3f235a574afd48a1f345bc2937dad6e7660648516c8029f4e76993ea74d.jpg
│ ├── iris-213cd179db580f8e633087dcda0969fd175d18d4f325cb5b4c5f394bbba0c1e0.jpg
│ ├── iris-21a1255e058722de1abe928e5bbe1c77bda31824c406c53f19530a3ca40be218.jpg
│ ├── iris-249370d38cc29bc2a4038e528f9c484c186fe46a126e4b6c76607860679c0453.jpg
│ ├── iris-2ac575a689662b7045c25e2554df5f985a3c6c0fd5236fabef8de9c78815330c.jpg
│ ├── iris-2c5b373c2a5fd214092eb578c75eb5dc84334e5f11a02f4fa23d5d316b18f770.jpg
│ ├── iris-2ecaad6dfe3d9b84a756bc2303a975a732718b954a6f54eae85f681ea3189b13.jpg
│ ├── iris-32827aec52e0f3fa131fa85f2092fc6fa02b1b80642740b59d029cef920c26b3.jpg
│ ├── iris-336fc3472b6465826f7cd87d5cef8f78d43cf2772ebe058ce71e1c5bad74c0e1.jpg
│ ├── iris-432026d8501abcd495bd98937a82213da97fca410af1c46889eabbcf2fd1b589.jpg
│ ├── iris-49a9158e46e788a39eeaefe82b19504d58dde167f540df6bc9492c3916d5f7ca.jpg
│ ├── iris-4b47f927405d90caa15cbf17b0442390fc71a2ca6fb8d07138e8de17d739e9a4.jpg
│ ├── iris-5691cad06fe37f743025c097fa9c4cec85e20ca3b0efff29175e60434e212421.jpg
│ ├── iris-5c38dba6f6c27064eb3920a5758e8f86c26fec662cc1ac4b5208d5f30d1e3ead.jpg
│ ├── iris-5da184e8620ebf0feef4d5ffe4346e6c44b2fb60cecc0320bd7726a1844b14cd.jpg
│ ├── iris-66eee9ff0bfa521905f733b2a0c6c5acad7b8f1a30d280ed4a17f54fe1822a7e.jpg
│ ├── iris-6815050b6117cf2e1fd60b1c33bfbb94837b8e173ff869f625757da4a04965c9.jpg
│ ├── iris-793fe85ddd6a97e9c9f184ed20d1d216e48bf85aa71633eff6d27073e0825d54.jpg
│ ├── iris-850229e6293a741277eb5efaa64d03c812f007c5d0f470992a8d4cfdb902230c.jpg
│ ├── iris-86d782d20ef7a60e905e367050b0413ca566acc672bc92add0bb0304faa54cfc.jpg
│ ├── iris-875a96790adc5672e044cf9da9d2edb397627884dfe91c488ab3fb65f65c80ff.jpg
│ ├── iris-96f06136df7a415550b90e443771d0b5b0cd990b503b64cc4987f5cb6797fa9b.jpg
│ ├── iris-9a889c96a37e8927f20773783a084f31897f075353d34a304c85e53be480e72a.jpg
│ ├── iris-9e3208f4f9fedc9598ddf26f77925a1e8df9d7865a4d6e5b4f74075d558d6a5e.jpg
│ ├── iris-a7e13b6f2d7f796768d898f5f66dceefdbd566dd4406eea9f266fc16dd68a6f2.jpg
│ ├── iris-b026efb61a9e3876749536afe183d2ace078e5e29615b07ac8792ab55ba90ebc.jpg
│ ├── iris-b3c086333cb5ccb7bb66a163cf4bf449dc0f28df27d6580a35832f32fd67bfc9.jpg
│ ├── iris-b795e034b6ea08d3cd9acaa434c67aca9d17016991e8dd7d6fd19ae8f6120b77.jpg
│ ├── iris-bb4a7ad4c844987bc9dc9dfad2b363698811efe3615512997a13cd191c23febc.jpg
│ ├── iris-bd60a6ed0369df4bea1934ef52277c32757838123456a595c0f2484959553a36.jpg
│ ├── iris-c15d6019ebe17d7446ced589ef5ef7a70474d35a8b072e0edfcec850b0a106db.jpg
│ ├── iris-c45295e76c6289504921412293d5ddbe4610bb6e3b593ea9ec90958e74b73ed2.jpg
│ ├── iris-c50d481f9fa3666c2c3808806c7c2945623f9d9a6a1d93a17133c4cb1560c41c.jpg
│ ├── iris-df4206653f1ec9909434323c05bb15ded18e72587e335f8905536c34a4be3d45.jpg
│ ├── iris-e45d869cb9d443b39d59e35c2f47870f5a2a335fce53f0c8a5bc615b9c53c429.jpg
│ ├── iris-e76fa5406e02a312c102f16eb5d27c7e0de37b35f801e1ed4c28bd4caf133e7a.jpg
│ ├── iris-e8d3fd862aae1c005bcc80a73fd34b9e683634933563e7538b520f26fd315478.jpg
│ ├── iris-ea578f650069a67e5e660bb22b46c23e0a182cbfb59cdf5448cf20ce858131b6.jpg
│ ├── iris-eba0c546e9b7b3d92f0b7eb98b2914810912990789479838807993d13787a2d9.jpg
│ ├── iris-f22d4b9605e62db13072246ff6925b9cf0240461f9dfc948d154b983db4243b9.jpg
│ ├── iris-fac5f8c23d8c50658db0f4e4a074c2f7771917eb52cbdf6eda50c12889510cf4.jpg
│ └── meta.csv
└── study2_raw_images/
├── iris-01cdd55ca6402713465841abddcce79a2e906e12edf95afb77c16bde4b4907dc.jpg
├── iris-02868b71ddd9b33ab795ac41609ea7b20a6e94f2543fad5d7fa11241d61feacf.jpg
├── iris-0415d2f3295db04bebc93249b685f7d7af7873faa911cd270ecd8363bd322ed5.jpg
├── iris-0c826b6f4648edf507e0cafdab53712bb6fd1f04dab453cee8db774a728dd640.jpg
├── iris-10fb9f154ead3c56ba0ab2c1ab609521c963f2326a648f82c9d7cabd178fc425.jpg
├── iris-14cbed88b0d2a929477bdf1299724f22d782e90f29ce55531f4a3d8608f7d926.jpg
├── iris-186fe29e32ee1405ddbdd36236dd7691a3c45ba78cc4c0bf11489fa09fbb1b65.jpg
├── iris-1b0b5aabd59e4c6ed1ceb54e57534d76f2f3f97e0a81800ff7ed901c35a424ab.jpg
├── iris-1d35672eb95f5b1cf14c2977eb025c246f83cdacd056115fdc93e946b56b610c.jpg
├── iris-1f941001f508ff1bd492457a90da64e52c461bfd64587a3cf7c6bf1bcb35adab.jpg
├── iris-2a09038b87009ecee5e5b4cd4cef068653809cc1e08984f193fad00f1c0df972.jpg
├── iris-308389e34b6d9a61828b339916aed7af295fdb1c7577c23fb37252937619e7e4.jpg
├── iris-30e4e56b1f170ff4863b178a0a43ea7a64fdd06c1f89a775ec4dbf5fec71e15c.jpg
├── iris-332953f4d6a355ca189e2508164b24360fc69f83304e7384ca2203ddcb7c73b5.jpg
├── iris-338fc323ed045a908fb1e8ff991255e1b8e01c967e36b054cb65edddf97b3bb0.jpg
├── iris-34a7cc16d26ba0883574e7a1c913ad50cf630e56ec08ee1113bf3584f4e40230.jpg
├── iris-360196ba36654c0d9070f95265a8a90bc224311eb34d1ab0cf851d8407d7c28e.jpg
├── iris-36132c6df6b47bda180b1daaafc7ac8a32fd7f9af83a92569da41429da49ea5b.jpg
├── iris-36f2b9282342292b67f38a55a62b0c66fa4e5bb58587f7fec90d1e93ea8c407a.jpg
├── iris-37ad07fd7b39bc377fa6e9cafdb6e0c57fb77df2c264fe631705a8436c0c2513.jpg
├── iris-3ba1625bb78e4b69b114bdafcdab64104b211d8ebadca89409e9e7ead6a0557c.jpg
├── iris-4c5d9a33327db025d9c391aeb182cbe20cfab4d4eb4ac951cc5cd15e132145d8.jpg
├── iris-522f3eb1807d015f99e66e73b19775800712890f2c7f5b777409a451fa47d532.jpg
├── iris-589fa96b9a3c2654cf08d05d3bebf4ab7bc23592d7d5a95218f9ff87612992fa.jpg
├── iris-61b71f1de04a03ce719094b65179b06e3cd80afa01622b30cda8c3e41de6bfaa.jpg
├── iris-62ef719cd70780088a4c140afae2a96c6ca9c22b72b078e3b9d25678d00b88a5.jpg
├── iris-819130af42335d4bb75bebb0d2ee2e353a89a3d518a1d2ce69842859c5668c5a.jpg
├── iris-8669e4937a2003054408afd228d99cb737e9db5088f42d292267c43a3889001a.jpg
├── iris-86c76e0f331bc62192c392cf7c3ea710d2272a8cc9928d2566a5fc4559e5dce4.jpg
├── iris-8a8bc54332a42bb35ee131d7b64e9375b4ac890632eb09e193835b838172d797.jpg
├── iris-8e9439ec7231fa3b9bc9f62a67af4e180466b32a72316600431b1ec93e63b296.jpg
├── iris-90b7d491b9a39bb5c8bb7649cce90ab7f483c2759fb55fda2d9067ac9eec7e39.jpg
├── iris-9dededf184993455c411a0ed81d6c3c55af7c610ccb55c6ae34dfac2f8bde978.jpg
├── iris-9e6ce91679c9aaceb3e9c930f11e788aacbfa8341a2a5737583c14a4d6666f3d.jpg
├── iris-a0e65269f7dc7801ac1ad8bd0c5aa547a70c7655447e921d1d4d153a9d23815e.jpg
├── iris-a445b0720254984275097c83afbdb1fe896cb010b5c662a6532ed0601ea24d7c.jpg
├── iris-a6b85bf1f3d18bbb6470440592834c2c7f081b490836392cf5f01636ee7cf658.jpg
├── iris-b005c82b844de575f0b972b9a1797b2b1fbe98c067c484a51006afc4f549ada4.jpg
├── iris-bfcf79b3b527eb64b78f9a068a1000042336e532f0f44e68f818dd13ab492a76.jpg
├── iris-c156236fb6e888764485e796f1f972bbc7ad960fe6330a7ce9182922046439c4.jpg
├── iris-d99d5fd2de5be1419cbd569570dbb6c9a6c8ec4f0a1ff5b55dc2607f6ecdca8f.jpg
├── iris-d9aae37a8fa6afdef2af170c266a597925eea935f4d070e979d565713ea62642.jpg
├── iris-dbc87fcecade2c070baaf99caf03f4f0f6e3aa977e34972383cb94d0efe8a95d.jpg
├── iris-e3d1a560d25cf573d2cbbf2fe6cd231819e998109a5cf1788d59fbb9859b3be2.jpg
├── iris-ec288bdad71388f907457db2476f12a5cb43c28cfa28d2a2077398a42b948a35.jpg
├── iris-ed5b4e072d43bc53a00a4a7f4d0f5d7c0cbd6a006e9c2d463128cedc956cb3de.jpg
├── iris-f3018a9440d17c265062d1c61475127f9952b6fe951d38fd7700402d706c0b01.jpg
├── iris-f47c5963cdbaa3238ba2d446848e8449c6af83e663f0a9216cf0baba8429b36f.jpg
├── iris-fa4b6d7e3617216104b1405cda21bf234840cd84a2c1966034caa63def2f64f0.jpg
├── iris-fc4b0cc65387ff78471659d14a78f0309a76f4c3ec641b871e40b40424255097.jpg
└── meta.csv
Let’s create an artifact for the first sub-folder.
artifact = ln.Artifact("s3://lamindata/iris_studies/study0_raw_images").save()
artifact
artifact <- ln$Artifact("s3://lamindata/iris_studies/study0_raw_images")$save()
artifact
Show code cell outputs
→ referenced read-only storage location at s3://lamindata, is managed by instance with uid 4XIuR0tvaiXM
Artifact(uid='0DukjvqoYOvWH7uI0000', version_tag=None, is_latest=True, key='iris_studies/study0_raw_images', description=None, suffix='', kind=None, otype=None, size=658465, hash='IVKGMfNwi8zKvnpaD_gG7w', n_files=51, n_observations=None, branch_id=1, created_on_id=1, space_id=1, storage_id=2, run_id=1, schema_id=None, created_by_id=1, created_at=2026-02-24 15:35:35 UTC, is_locked=False)
As you see from path, the folder was merely registered in its present storage location without copying it.
artifact.path
artifact$path
Show code cell outputs
S3QueryPath('s3://lamindata/iris_studies/study0_raw_images')
LaminDB keeps track of all your storage locations.
ln.Storage.to_dataframe()
ln$Storage$to_dataframe()
Show code cell outputs
| uid | root | description | type | region | instance_uid | is_locked | created_at | branch_id | created_on_id | space_id | created_by_id | run_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||
| 2 | YmV3ZoHvAAAA | s3://lamindata | None | s3 | us-east-1 | 4XIuR0tvaiXM | False | 2026-02-24 15:35:35.782000+00:00 | 1 | 1 | 1 | 1 | 1.0 |
| 1 | VLuWYwO8Gpy1 | /home/runner/work/lamin-docs/lamin-docs/docs/l... | None | local | None | 5WCoFhciyw3n | False | 2026-02-24 15:35:21.870000+00:00 | 1 | 1 | 1 | 1 | NaN |
How do I update or delete an artifact?
artifact.description = "My new description" # change description
artifact.save() # save the change to the database
artifact.delete() # move to trash
artifact.delete(permanent=True) # permanently delete
How do I create an artifact for a local file or folder?
Source path is local:
ln.Artifact("./my_data.fcs", key="my_dataset.fcs")
ln.Artifact("./my_images/", key="my_images")
Upon artifact.save(), the source path will be copied or uploaded into your instance’s current storage, visible & changeable via ln.settings.storage.
If the source path is remote or already in a registered storage location (one that’s registered in ln.Storage), artifact.save() will not trigger a copy or upload but register the existing path.
ln.Artifact("s3://my-bucket/my_dataset.fcs") # key is auto-populated from S3, you can optionally pass a description
ln.Artifact("s3://my-bucket/my_images/") # key is auto-populated from S3, you can optionally pass a description
You can use any storage location supported by `fsspec`.
Which fields are populated when creating an artifact record?
Basic fields:
uid: universal IDkey: a (virtual) relative path of the artifact instoragedescription: an optional string descriptionstorage: the storage location (the root, say, an S3 bucket or a local directory)suffix: an optional file/path suffixsize: the artifact size in byteshash: a hash useful to check for integrity and collisions (is this artifact already stored?)hash_type: the type of the hashcreated_at: time of creationupdated_at: time of last update
Provenance-related fields:
created_by: theUserwho created the artifact
For a full reference, see Artifact.
What exactly happens during save?
In the database: An artifact record is inserted into the Artifact registry. If the artifact record exists already, it’s returned.
In storage:
If the default storage is in the cloud,
.save()triggers an upload for a local artifact.If the artifact is already in a registered storage location, only the metadata of the record is saved to the
artifactregistry.
How does LaminDB compare to a AWS S3?
LaminDB provides a database on top of AWS S3 (or GCP storage, file systems, etc.).
Similar to organizing files with paths, you can organize artifacts using the key parameter of Artifact.
However, you’ll see that you can more conveniently query data by entities you care about: people, code, experiments, genes, proteins, cell types, etc.
Are artifacts aware of array-like data?
Yes.
You can make artifacts from paths referencing array-like objects:
ln.Artifact("./my_anndata.h5ad", key="my_anndata.h5ad")
ln.Artifact("./my_zarr_array/", key="my_zarr_array")
Or from in-memory objects:
ln.Artifact.from_dataframe(df, key="my_dataframe.parquet")
ln.Artifact.from_anndata(adata, key="my_anndata.h5ad")
You can open large artifacts for slicing from the cloud or load small artifacts directly into memory via:
artifact.open()
Manage biological registries
Every bionty registry is based on configurable public ontologies (>20 of them) that are automatically leveraged during validation & annotation. Sometimes you want to access the public ontology directly.
import bionty as bt
cell_type_ontology = bt.CellType.public()
cell_type_ontology
bt <- import_module("bionty")
cell_type_ontology <- bt$CellType$public()
cell_type_ontology
Show code cell outputs
PublicOntology
Entity: CellType
Organism: all
Source: cl, 2025-12-17
#terms: 3437
The returned object can be searched like you can search a registry.
cell_type_ontology.search("gamma-delta T cell").head(2)
cell_type_ontology$search("gamma-delta T cell")$head(2)
Show code cell outputs
| name | definition | synonyms | parents | |
|---|---|---|---|---|
| ontology_id | ||||
| CL:0000798 | gamma-delta T cell | A T Cell That Expresses A Gamma-Delta T Cell R... | gammadelta T cell|gamma-delta T-lymphocyte|gam... | [CL:0000084] |
| CL:4033072 | cycling gamma-delta T cell | A(N) Gamma-Delta T Cell That Is Cycling. | proliferating gamma-delta T cell | [CL:0000798, CL:4033069] |
Because you can’t update an external public ontology, you update the content of the corresponding registry. Here, you create a new cell type.
# create an ontology-coupled cell type record and save it
neuron = bt.CellType.from_source(name="neuron").save()
# create a record to track a new cell state
new_cell_state = bt.CellType(
name="my neuron cell state", description="explains X"
).save()
# express that it's a neuron state
new_cell_state.parents.add(neuron)
# view ontological hierarchy
new_cell_state.view_parents(distance=2)
# create an ontology-coupled cell type record and save it
neuron <- bt$CellType$from_source(name = "neuron")$save()
# create a record to track a new cell state
new_cell_state <- bt$CellType(
name = "my neuron cell state", description = "explains X"
)$save()
# express that it's a neuron state
new_cell_state$parents$add(neuron)
# view ontological hierarchy
new_cell_state$view_parents(distance = 2)
Show code cell outputs
Manage AnnData objects¶
LaminDB supports a growing number of data structures: DataFrame, AnnData, MuData, SpatialData, and Tiledbsoma with their corresponding representations in storage.
Let’s go through the example of the quickstart, but store the dataset in an AnnData this time.
# define var schema
var_schema = ln.Schema(itype=bt.Gene.ensembl_gene_id, dtype=int).save()
# define composite schema
anndata_schema = ln.Schema(
otype="AnnData", slots={"obs": schema, "var.T": var_schema}
).save()
# define var schema
var_schema <- ln$Schema(itype = bt$Gene$ensembl_gene_id, dtype = "int")$save()
# define composite schema
anndata_schema <- ln$Schema(
otype = "AnnData", slots = list(obs = schema, var$T = var_schema)
)$save()
Validate & annotate an AnnData.
import anndata as ad
# store the dataset as an AnnData object to distinguish data from metadata
adata = ad.AnnData(df.iloc[:, :3], obs=df.iloc[:, 3:-1])
# save curated artifact
artifact = ln.Artifact.from_anndata(
adata, key="my_datasets/my_rnaseq1.h5ad", schema=anndata_schema
).save()
artifact.describe()
ad <- import_module("anndata")
# store the dataset as an AnnData object to distinguish data from metadata
adata <- anndata::AnnData(df$iloclist(:, :3), obs = df$iloclist(:, 3:-1))
# save curated artifact
artifact <- ln$Artifact$from_anndata(
adata, key = "my_datasets/my_rnaseq1.h5ad", schema = anndata_schema
)$save()
artifact$describe()
Show code cell outputs
→ loading artifact into memory for validation
→ returning schema with same hash: Schema(uid='5we5Kvp3E9AayEr7', is_type=False, name=None, description=None, n_members=8, coerce=None, flexible=False, itype='Feature', otype=None, hash='V3yLoat04fZMXmIp8VmHxw', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, created_on_id=1, space_id=1, created_by_id=1, run_id=1, type_id=None, created_at=2026-02-24 15:35:32 UTC, is_locked=False)
Artifact: my_datasets/my_rnaseq1.h5ad (0000) ├── uid: ZdvHx4tIkl3BFSBa0000 run: dHStEMK (tutorial.ipynb) │ kind: dataset otype: AnnData │ hash: TWUlDi-V-Try1uCxWJ65nQ size: 29.5 KB │ branch: main space: all │ created_at: 2026-02-24 15:35:40 UTC created_by: anonymous │ n_observations: 3 ├── storage/path: /home/runner/work/lamin-docs/lamin-docs/docs/lamindb-tutorial/.lamindb/ZdvHx4tIkl3BFSBa0000.h5ad ├── Dataset features │ ├── obs (8) │ │ assay_oid bionty.ExperimentalFactor.ontology… EFO:0008913 │ │ cell_type_by_expert bionty.CellType B cell, CD8-positive, alpha-beta T cell │ │ cell_type_by_model bionty.CellType B cell, T cell │ │ concentration str │ │ donor str │ │ perturbation Record DMSO, IFNG │ │ sample_note str │ │ treatment_time_h num │ └── var.T (3 bionty.Gene.ensembl… │ CD14 num │ CD4 num │ CD8A num └── Labels └── .records Record DMSO, IFNG .cell_types bionty.CellType T cell, B cell, CD8-positive, alpha-be… .experimental_factors bionty.ExperimentalFactor single-cell RNA sequencing
Because AnnData separates the high-dimensional count matrix that’s typically indexed with Ensembl gene ids from the metadata, we’re now working with two types of feature sets (bt.Gene for the counts and ln.Feature for the metadata). These correspond to the obs and the var schema in the anndata_schema.
If you want to find a dataset by whether it measured CD8A, you can do so as as follows.
# query for all feature sets that contain CD8A
feature_sets = ln.Schema.filter(genes__symbol="CD8A").all()
# query for all artifacts linked to these feature sets
ln.Artifact.filter(feature_sets__in=feature_sets).to_dataframe()
# query for all feature sets that contain CD8A
feature_sets <- ln$Schema$filter(genes__symbol = "CD8A")$all()
# query for all artifacts linked to these feature sets
ln$Artifact$filter(feature_sets__in = feature_sets)$to_dataframe()
Show code cell outputs
| uid | key | description | suffix | kind | otype | size | hash | n_files | n_observations | ... | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | storage_id | run_id | schema_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 4 | ZdvHx4tIkl3BFSBa0000 | my_datasets/my_rnaseq1.h5ad | None | .h5ad | dataset | AnnData | 30232 | TWUlDi-V-Try1uCxWJ65nQ | None | 3 | ... | True | False | 2026-02-24 15:35:40.377000+00:00 | 1 | 1 | 1 | 1 | 1 | 4 | 1 |
1 rows × 21 columns
Scale learning¶
How do you integrate new datasets with your existing datasets? Leverage Collection.
# a new dataset
df2 = ln.examples.datasets.mini_immuno.get_dataset2(otype="DataFrame")
adata = ad.AnnData(df2.iloc[:, :3], obs=df2.iloc[:, 3:-1])
artifact2 = ln.Artifact.from_anndata(
adata, key="my_datasets/my_rnaseq2.h5ad", schema=anndata_schema
).save()
# a new dataset
df2 <- ln$examples$datasets$mini_immuno$get_dataset2(otype = "DataFrame")
adata <- anndata::AnnData(df2$iloclist(:, :3), obs = df2$iloclist(:, 3:-1))
artifact2 <- ln$Artifact$from_anndata(
adata, key = "my_datasets/my_rnaseq2.h5ad", schema = anndata_schema
)$save()
Show code cell outputs
→ loading artifact into memory for validation
Create a collection using Collection.
collection = ln.Collection([artifact, artifact2], key="my-RNA-seq-collection").save()
collection.describe()
collection.view_lineage()
collection <- ln$Collection(list(artifact, artifact2), key = "my-RNA-seq-collection")$save()
collection$describe()
collection$view_lineage()
Show code cell outputs
Collection: my-RNA-seq-collection (0000) └── uid: HcfSoAZ1GQ8gM9nt0000 run: dHStEMK (tutorial.ipynb) branch: main space: all created_at: 2026-02-24 15:35:42 UTC created_by: anonymous
# if it's small enough, you can load the entire collection into memory as if it was one
collection.load()
# typically, it's too big, hence, open it for streaming (if the backend allows it)
# collection.open()
# or iterate over its artifacts
collection.artifacts.all()
# or look at a DataFrame listing the artifacts
collection.artifacts.to_dataframe()
# if it's small enough, you can load the entire collection into memory as if it was one
collection$load()
# typically, it's too big, hence, open it for streaming (if the backend allows it)
# collection$open()
# or iterate over its artifacts
collection$artifacts$all()
# or look at a DataFrame listing the artifacts
collection$artifacts$to_dataframe()
Show code cell outputs
| uid | key | description | suffix | kind | otype | size | hash | n_files | n_observations | ... | is_latest | is_locked | created_at | branch_id | created_on_id | space_id | storage_id | run_id | schema_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 5 | 59oAbVdOaYIIIBt80000 | my_datasets/my_rnaseq2.h5ad | None | .h5ad | dataset | AnnData | 23712 | evwa9JaetcfvvMASeqqDVA | None | 3 | ... | True | False | 2026-02-24 15:35:42.740000+00:00 | 1 | 1 | 1 | 1 | 1 | 4 | 1 |
| 4 | ZdvHx4tIkl3BFSBa0000 | my_datasets/my_rnaseq1.h5ad | None | .h5ad | dataset | AnnData | 30232 | TWUlDi-V-Try1uCxWJ65nQ | None | 3 | ... | True | False | 2026-02-24 15:35:40.377000+00:00 | 1 | 1 | 1 | 1 | 1 | 4 | 1 |
2 rows × 21 columns
Directly train models on collections of AnnData.
# to train models, batch iterate through the collection as if it was one array
from torch.utils.data import DataLoader, WeightedRandomSampler
dataset = collection.mapped(obs_keys=["cell_medium"])
sampler = WeightedRandomSampler(
weights=dataset.get_label_weights("cell_medium"), num_samples=len(dataset)
)
data_loader = DataLoader(dataset, batch_size=2, sampler=sampler)
for batch in data_loader:
pass
Read this blog post for more on training models on distributed datasets.