Vitessce: SpatialData 
 ¶
¶
This tutorial demonstrates how to create interactive Vitessce visualizations for SpatialData objects stored as LaminDB artifacts. It requires a remote LaminDB instance with cloud storage to enable the Vitessce button (shown below) in the web interface.

For visualizing AnnData objects, see the Vitessce: AnnData guide.
We’ll work with spatial transcriptomics data in Zarr, OME-TIFF, and OME-Zarr formats.
# pip install 'vitessce[all]>=3.5.0' lamindb
!lamin connect laminlabs/lamindata # <-- replace with your remote instance
Show code cell output
→ connected lamindb: laminlabs/lamindata
• to map a local dev directory, call: lamin settings set dev-dir .
import vitessce as vit
import lamindb as ln
ln.track()
Show code cell output
→ connected lamindb: laminlabs/lamindata
→ found notebook vitessce2.ipynb, making new version -- anticipating changes
→ created Transform('yslqNnvM5Gzh000B', key='vitessce2.ipynb'), started new Run('TEVJt8sT3Mu0a0uZ') at 2026-02-24 14:51:21 UTC
→ notebook imports: lamindb==2.2.1 vitessce==3.8.2
• recommendation: to identify the notebook across renames, pass the uid: ln.track("yslqNnvM5Gzh")
Visualize a SpatialData object (Zarr format)¶
Here we use an example Visium dataset that has been previously ingested into the public laminlabs/lamindata instance in this transform.
sdata_zarr_artifact = ln.Artifact.get(key="vitessce_examples/visium.sdata.zarr")
Save a VitessceConfig object¶
You can create a dashboard for one or several datasets by using Vitessce’s component API.
Here, we use the SpatialDataWrapper class to specify which parts of the SpatialData object will be loaded for visualization.
vc = vit.VitessceConfig(
schema_version="1.0.18",
description=sdata_zarr_artifact.description,
)
dataset_uid = "sdata_visium"
dataset = vc.add_dataset(name="Breast Cancer Visium", uid=dataset_uid).add_object(
vit.SpatialDataWrapper(
sdata_artifact=sdata_zarr_artifact,
# The following paths are relative to the root of the SpatialData zarr store on-disk.
image_path="images/CytAssist_FFPE_Human_Breast_Cancer_full_image",
table_path="tables/table",
obs_feature_matrix_path="tables/table/X",
obs_spots_path="shapes/CytAssist_FFPE_Human_Breast_Cancer",
region="CytAssist_FFPE_Human_Breast_Cancer",
coordinate_system="global",
coordination_values={
# The following tells Vitessce to consider each observation as a "spot"
"obsType": "spot",
},
)
)
# Add views (visualizations) to the configuration:
spatial = vc.add_view("spatialBeta", dataset=dataset)
feature_list = vc.add_view("featureList", dataset=dataset)
layer_controller = vc.add_view("layerControllerBeta", dataset=dataset)
# Initialize visual properties for multiple linked views:
vc.link_views_by_dict(
[spatial, layer_controller],
{
"imageLayer": vit.CoordinationLevel(
[
{
"photometricInterpretation": "RGB",
}
]
),
},
scope_prefix=vit.get_initial_coordination_scope_prefix(dataset_uid, "image"),
)
vc.link_views([spatial, layer_controller, feature_list], ["obsType"], ["spot"])
# Layout the views
vc.layout(spatial | (feature_list / layer_controller));
Save the VitessceConfig object.
sdata_vc_artifact = ln.integrations.save_vitessce_config(
vc,
description="View Visium SpatialData Example in Vitessce",
)
Show code cell output
→ VitessceConfig references these artifacts:
Artifact(uid='Z1Q9alE7dBr5lZty0000', version_tag=None, is_latest=True, key='vitessce_examples/visium.sdata.zarr', description='Visium SpatialData Example', suffix='.zarr', kind='dataset', otype='SpatialData', size=1473181518, hash='iDvq8-aqbbY8EBA1QCoMzw', n_files=9170, n_observations=None, branch_id=1, created_on_id=1, space_id=1, storage_id=2, run_id=1933, schema_id=None, created_by_id=40, created_at=2026-01-20 19:53:35 UTC, is_locked=False)
→ returning artifact with same hash: Artifact(uid='CsXv773YgZkfBSfx0000', version_tag=None, is_latest=True, key=None, description='View Visium SpatialData Example in Vitessce', suffix='.vitessce.json', kind='__lamindb_config__', otype=None, size=1810, hash='gbTMZ_T-ecR3A-8XbjO3RQ', n_files=None, n_observations=None, branch_id=1, created_on_id=1, space_id=1, storage_id=2, run_id=1945, schema_id=None, created_by_id=18, created_at=2026-01-20 20:45:29 UTC, is_locked=False); to track this artifact as an input, use: ln.Artifact.get()
→ VitessceConfig: https://lamin.ai/laminlabs/lamindata/artifact/CsXv773YgZkfBSfx0000
→ Dataset: https://lamin.ai/laminlabs/lamindata/artifact/Z1Q9alE7dBr5lZty0000
Note
After running save_vitessce_config, a Vitessce button will appear next to the dataset on the Artifacts or Collections page of the web interface.
If your VitessceConfig object references data from multiple artifacts, the Vitessce button will appear next to a collection that groups these artifacts (on the Collections tab of the Artifacts page).
Clicking the Vitessce button for this artifact launches an interactive viewer with spatial coordinates, gene expression, and tissue image:
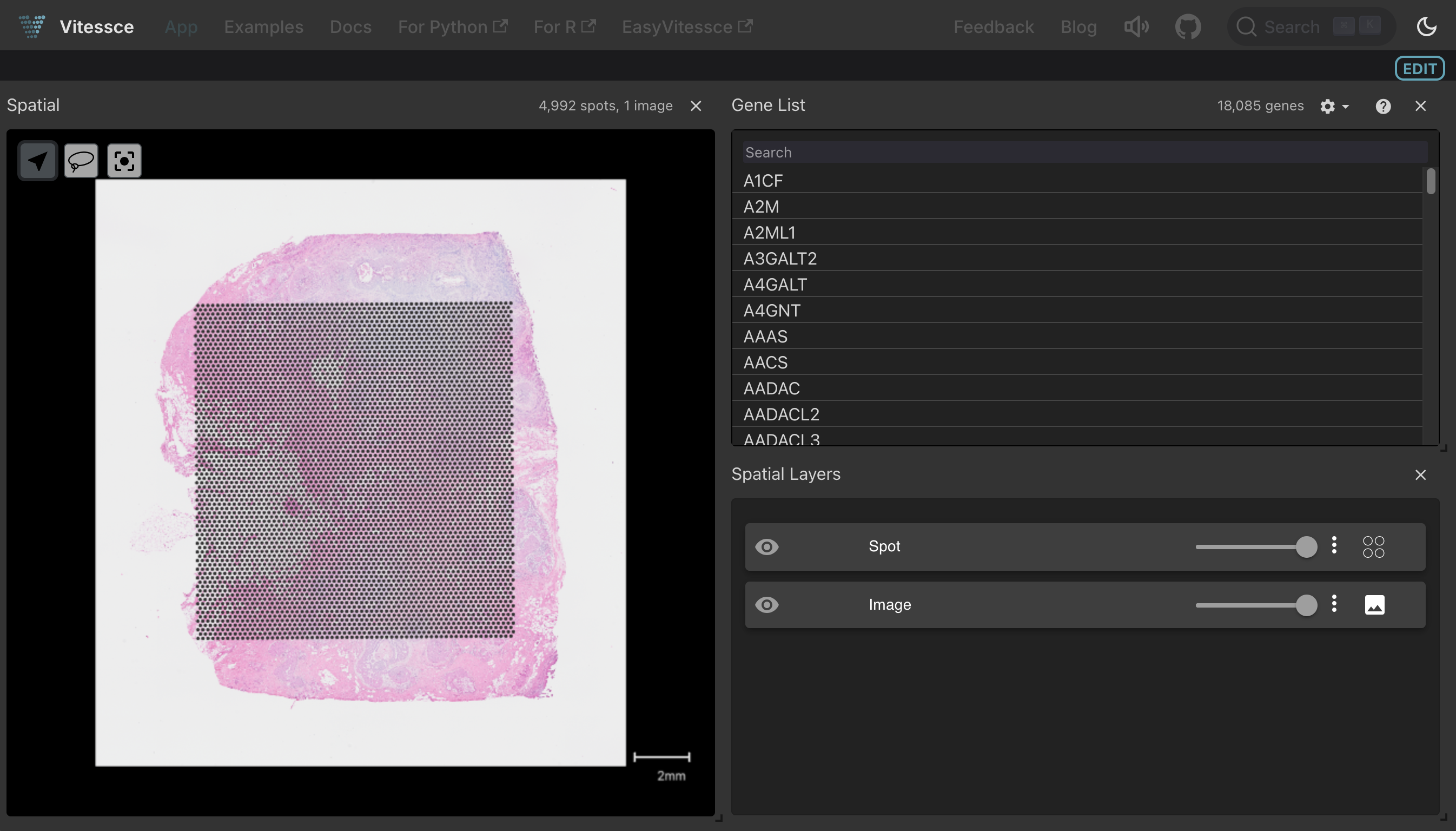
Visualize an image (OME-TIFF format)¶
Vitesse can visualize data from multiple bioimaging file formats, including OME-TIFF.
Again, we use an example dataset that was previously ingested into the laminlabs/lamindata instance in this transform.
ome_tiff_artifact = ln.Artifact.get(
key="vitessce_examples/VAN0006-LK-2-85-PAS_registered.ome.tif"
)
When using OME-TIFF files, we can use generate-tiff-offsets to create an index for the bytes within the OME-TIFF file.
We store these to a companion .offsets.json file, making it more efficient to load image subsets.
We use a file generated by the transform above.
offsets_artifact = ln.Artifact.get(
key="vitessce_examples/VAN0006-LK-2-85-PAS_registered.offsets.json"
)
Save a VitessceConfig object¶
You can create a dashboard for one or several datasets by using Vitessce’s component API.
Here, we use the ImageOmeTiffWrapper class to specify which pair of OME-TIFF file and offsets JSON file to load.
vc = vit.VitessceConfig(
schema_version="1.0.18", description=ome_tiff_artifact.description
)
dataset = vc.add_dataset("Image").add_object(
vit.ImageOmeTiffWrapper(
img_artifact=ome_tiff_artifact,
offsets_artifact=offsets_artifact,
)
)
spatial = vc.add_view("spatialBeta", dataset=dataset)
layer_controller = vc.add_view("layerControllerBeta", dataset=dataset)
vc.layout(spatial | layer_controller);
Save the VitessceConfig object.
ome_tiff_vc_artifact = ln.integrations.save_vitessce_config(
vc,
description="View PAS OME-TIFF, Neumann et al., 2020 in Vitessce",
)
Show code cell output
→ VitessceConfig references these artifacts:
Artifact(uid='wkRgbeUNJLAPN9Re0000', version_tag=None, is_latest=True, key='vitessce_examples/VAN0006-LK-2-85-PAS_registered.ome.tif', description='PAS OME-TIFF file, Neumann et al., 2020', suffix='.tif', kind='dataset', otype=None, size=1021561156, hash='pa3FYqKL6afG8PduprhRJk', n_files=None, n_observations=None, branch_id=1, created_on_id=1, space_id=1, storage_id=2, run_id=2244, schema_id=None, created_by_id=40, created_at=2026-01-28 23:32:35 UTC, is_locked=False)
Artifact(uid='mPRpRQxVjW5UxKKz0000', version_tag=None, is_latest=True, key='vitessce_examples/VAN0006-LK-2-85-PAS_registered.offsets.json', description='PAS offsets file, Neumann et al., 2020', suffix='.json', kind=None, otype=None, size=36, hash='5ADv5d7DI5oYVaMOPnhqiw', n_files=None, n_observations=None, branch_id=1, created_on_id=1, space_id=1, storage_id=2, run_id=2244, schema_id=None, created_by_id=40, created_at=2026-01-28 23:36:27 UTC, is_locked=False)
! returning collection with same hash: Collection(uid='DMF53hCJZQdnVIzT0000', version_tag=None, is_latest=True, key='View PAS OME-TIFF, Neumann et al., 2020 in Vitessce', description=None, hash='vpleElXxZiaZblQ09q9K-g', reference=None, reference_type=None, meta_artifact=None, branch_id=1, created_on_id=1, space_id=1, created_by_id=18, run_id=2250, created_at=2026-01-28 23:40:21 UTC, is_locked=False); if you intended to query to track this collection as an input, use: ln.Collection.get()
→ returning artifact with same hash: Artifact(uid='x6OfjnJaOTtTFPhW0000', version_tag=None, is_latest=True, key=None, description='View PAS OME-TIFF, Neumann et al., 2020 in Vitessce', suffix='.vitessce.json', kind='__lamindb_config__', otype=None, size=654, hash='fKTNFVp7GP_UEdEcnIaHvA', n_files=None, n_observations=None, branch_id=1, created_on_id=1, space_id=1, storage_id=2, run_id=2260, schema_id=None, created_by_id=18, created_at=2026-01-28 23:40:22 UTC, is_locked=False); to track this artifact as an input, use: ln.Artifact.get()
→ VitessceConfig: https://lamin.ai/laminlabs/lamindata/artifact/x6OfjnJaOTtTFPhW0000
→ Collection: https://lamin.ai/laminlabs/lamindata/collection/DMF53hCJZQdnVIzT0000
Note
After running save_vitessce_config, a Vitessce button will appear next to the dataset on the Artifacts or Collections page of the web interface.
If your VitessceConfig object references data from multiple artifacts, the Vitessce button will appear next to a collection that groups these artifacts (on the Collections tab of the Artifacts page).
In the case of OME-TIFF, the presence of the corresponding offsets JSON file will result in the creation of a collection.
Clicking the Vitessce button for this collection launches an interactive tissue image viewer:
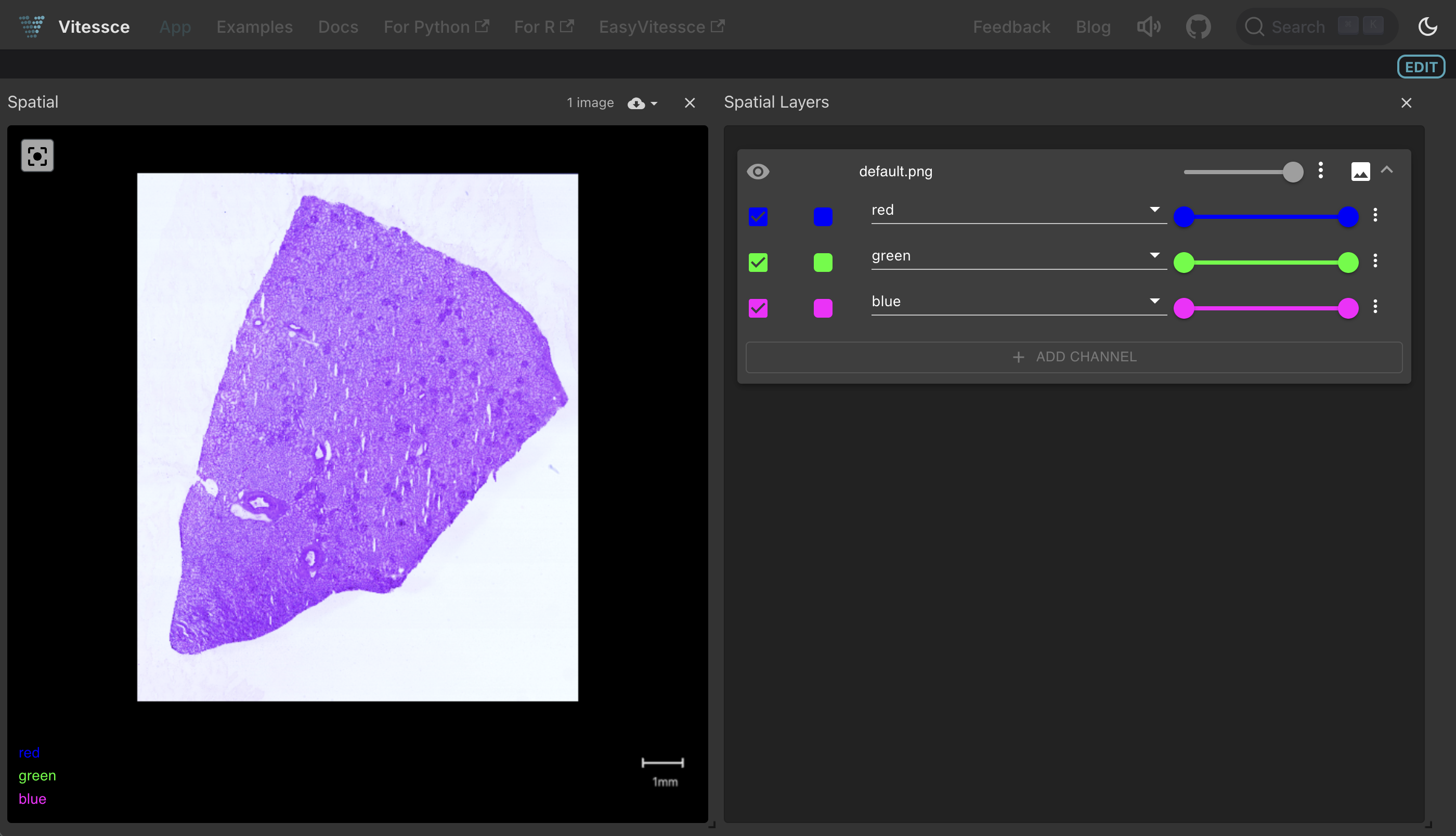
Visualize an image (OME-Zarr format)¶
We retrieve an OME-Zarr (also known as OME-NGFF) formatted image that was ingested into laminlabs/lamindata alongside the previous datasets.
ome_zarr_artifact = ln.Artifact.get(key="vitessce_examples/visium.ome.zarr")
Save a VitessceConfig object¶
You can create a dashboard for one or several datasets by using Vitessce’s component API. Here, we use the ImageOmeZarrWrapper class to specify an OME-Zarr file to load for visualization.
vc = vit.VitessceConfig(
schema_version="1.0.18", description=ome_zarr_artifact.description
)
dataset_uid = "ome_zarr_image"
dataset = vc.add_dataset("Image", uid=dataset_uid).add_object(
vit.ImageOmeZarrWrapper(
img_artifact=ome_zarr_artifact,
)
)
spatial = vc.add_view("spatialBeta", dataset=dataset)
layer_controller = vc.add_view("layerControllerBeta", dataset=dataset)
vc.link_views_by_dict(
[spatial, layer_controller],
{
"imageLayer": vit.CoordinationLevel(
[
{
"photometricInterpretation": "RGB",
}
]
),
},
scope_prefix=vit.get_initial_coordination_scope_prefix(dataset_uid, "image"),
)
vc.layout(spatial | layer_controller);
Save the VitessceConfig object.
ome_zarr_vc_artifact = ln.integrations.save_vitessce_config(
vc,
description="View Visium OME-Zarr Example in Vitessce",
)
Show code cell output
→ VitessceConfig references these artifacts:
Artifact(uid='dYRJon1HZoJuWhz80000', version_tag=None, is_latest=True, key='vitessce_examples/visium.ome.zarr', description='Visium OME-Zarr Example', suffix='.ome.zarr', kind='dataset', otype='SpatialData', size=1400251884, hash='LNdpQjV21yjPlL8hgJW6dA', n_files=8809, n_observations=None, branch_id=1, created_on_id=1, space_id=1, storage_id=2, run_id=1933, schema_id=None, created_by_id=40, created_at=2026-01-20 20:00:29 UTC, is_locked=False)
→ returning artifact with same hash: Artifact(uid='lcsFUjaeTpIAfzN40000', version_tag=None, is_latest=True, key=None, description='View Visium OME-Zarr Example in Vitessce', suffix='.vitessce.json', kind='__lamindb_config__', otype=None, size=1214, hash='zqAaftDfeoUzR0rftubZWA', n_files=None, n_observations=None, branch_id=1, created_on_id=1, space_id=1, storage_id=2, run_id=1947, schema_id=None, created_by_id=18, created_at=2026-01-20 20:45:31 UTC, is_locked=False); to track this artifact as an input, use: ln.Artifact.get()
→ VitessceConfig: https://lamin.ai/laminlabs/lamindata/artifact/lcsFUjaeTpIAfzN40000
→ Dataset: https://lamin.ai/laminlabs/lamindata/artifact/dYRJon1HZoJuWhz80000
Note
After running save_vitessce_config, a Vitessce button will appear next to the dataset on the Artifacts or Collections page of the web interface.
If your VitessceConfig object references data from multiple artifacts, the Vitessce button will appear next to a collection that groups these artifacts (on the Collections tab of the Artifacts page).
Clicking the Vitessce button for this artifact launches an interactive tissue image viewer:
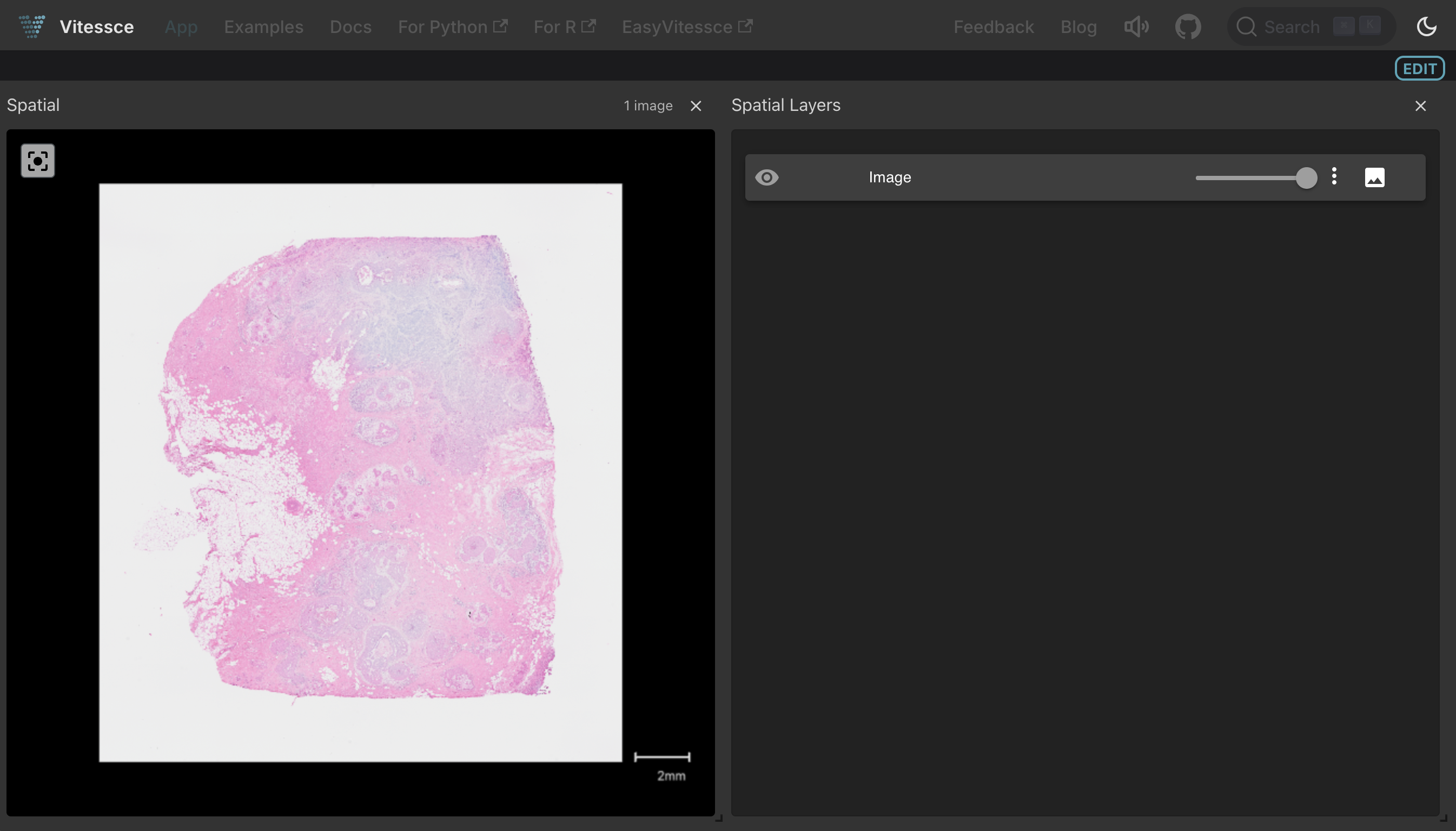
Show code cell content
# compare the generated vitessce config to the public one (SpatialData) on vitessce/examples
db = ln.DB("vitessce/examples")
public_vc_json = db.Artifact.get("Xot2a5ZAcTW3fClG0000").load()
sdata_vc_json = sdata_vc_artifact.load()
assert public_vc_json["layout"] == sdata_vc_json["layout"]
assert public_vc_json["coordinationSpace"] == sdata_vc_json["coordinationSpace"]
# compare the generated vitessce config json to the public one (OME-TIFF) on vitessce/examples
public_vc_json = db.Artifact.get("QtF1OEtYyUe1EQ1k0000").load()
ome_tiff_vc_json = ome_tiff_vc_artifact.load()
assert public_vc_json["layout"] == ome_tiff_vc_json["layout"]
assert public_vc_json["coordinationSpace"] == ome_tiff_vc_json["coordinationSpace"]
# compare the generated vitessce config to the public one (OME-Zarr) on vitessce/examples
public_vc_json = db.Artifact.get("cjvX6EFrdSwsxOQl0000").load()
ome_zarr_vc_json = ome_zarr_vc_artifact.load()
assert public_vc_json["layout"] == ome_zarr_vc_json["layout"]
assert public_vc_json["coordinationSpace"] == ome_zarr_vc_json["coordinationSpace"]
→ mapped: Artifact(uid='Xot2a5ZAcTW3fClG0000')
→ mapped: Artifact(uid='QtF1OEtYyUe1EQ1k0000')
→ mapped: Artifact(uid='cjvX6EFrdSwsxOQl0000')
ln.finish()
Show code cell output
! returning transform with same hash & key: Transform(uid='yslqNnvM5Gzh000A', version_tag=None, is_latest=False, key='vitessce2.ipynb', description='Vitessce: SpatialData [](https://github.com/laminlabs/lamin-spatial/blob/main/docs/vitessce2.md)', kind='notebook', hash='XqNogK20E9syLdG8wMo58A', reference=None, reference_type=None, environment=None, plan=None, branch_id=1, created_on_id=1, space_id=1, created_by_id=18, created_at=2026-02-05 15:41:18 UTC, is_locked=False)
• new latest Transform version is: yslqNnvM5Gzh000A
→ finished Run('TEVJt8sT3Mu0a0uZ') after 9s at 2026-02-24 14:51:30 UTC
→ go to: https://lamin.ai/laminlabs/lamindata/transform/yslqNnvM5Gzh000A
→ to update your notebook from the CLI, run: lamin save /home/runner/work/lamin-spatial/lamin-spatial/docs/vitessce2.ipynb